The Ekaterina Heldwein Lab
Viral egress: how do herpesviruses escape out of the infected cell?
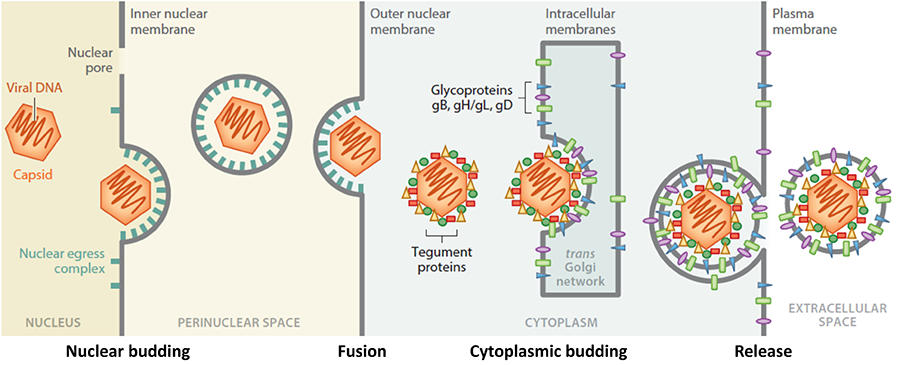
During herpesvirus egress, nascent capsids traverse multiple membranes, eventually forming mature infectious virions that exit the cell. Although egress is a process used by many viruses, unlike most other viruses, herpesviruses bud twice: first, at the nuclear envelope and then at the cytoplasmic membranes. Herpesviruses are the only known animal viruses that bud at the nuclear envelope. This unusual nuclear budding allows the viral capsid to escape from the nucleus into the cytoplasm. Herpesviral capsids are assembled in the nucleus but are too large to exit the nucleus through the nuclear pores. So, instead, they bud at the inner nuclear membrane (INM) forming perinuclear viral particles that pinch off into the perinuclear space (a process termed primary envelopment) and subsequently fuse with the outer nuclear membrane (ONM) (a process termed de-envelopment). The resulting cytoplasmic capsids bud again at the membranes derived from the trans-Golgi network (a process termed secondary envelopment), which generates mature virions that exit the cell by exocytosis. Egress is orchestrated by a complex network of protein-protein interactions. Although key participants have been identified, the knowledge of their specific roles and mechanisms is lacking. We will elucidate the structure-based mechanisms of nuclear budding, de-envelopment, and cytoplasmic budding by using a combination of structural biology, biochemistry, in-vitro reconstitution, and single-particle tracking assays. Our long-term goals are to elucidate the egress mechanisms at atomic-level detail and apply this knowledge to antiviral drug design.
Figure 1. Stages in herpesviral egress. Herpesviruses bud twice yet acquire only one envelope. Our work is focused on nuclear budding, tegument proteins, and cytoplasmic budding.
Viral invasion: how do herpesviruses get into host cells?
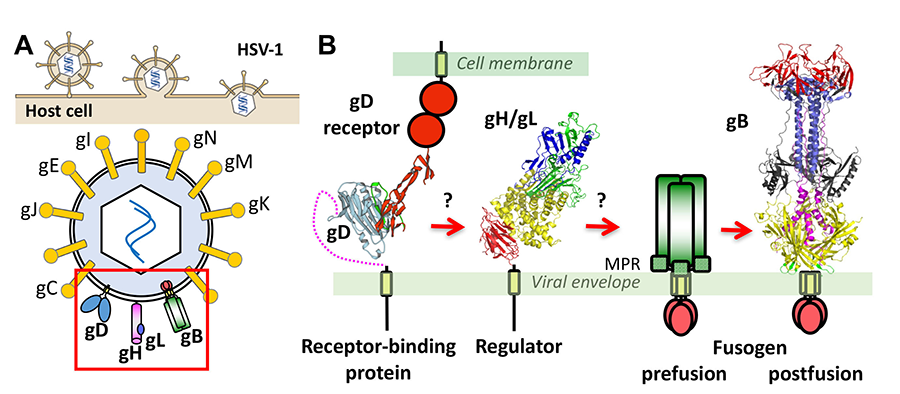
Herpesviruses enter cells by fusing their envelopes with the host membrane. While most enveloped viruses use a single viral protein, all herpesviruses inexplicably require multiple proteins. We have determined the structures of key herpesvirus penetration proteins, gB and gH/gL complex, and identified gB as the fusogen and gH/gL as a uniquely folded regulator. These findings upended the established way of thinking about the functions of these proteins and enabled a mechanistic exploration of herpesvirus entry. However, many aspects of herpesvirus-mediated fusion and entry remain undefined. What does gB look like in its active, prefusion form? How does gB catalyze membrane fusion? How is it triggered by other glycoproteins? How do multiple herpesviral glycoproteins work together to bring about membrane fusion? Answering these questions may explain the unique features of the herpesvirus entry machinery. In addition to dissecting the membrane fusion mechanism of HSV-1, we also strive to understand how and why herpesviruses choose different cell entry routes depending on the host cell type. We will use a combination of structural biology, biophysics, and cell biology to obtain answers to these and other outstanding questions in the field of herpesvirus entry. The proposed studies will provide fundamental structural and mechanistic insights into gB-mediated membrane fusion and entry that are currently lacking and may challenge some of the existing paradigms.
Figure 2. (A) Entry of HSV-1 into host cells occurs by membrane fusion and requires 4 viral proteins and a cell receptor. (B) Current model proposes that upon binding receptor, gD changes its conformation, which signals to gH/gL, which activates gB, the membrane fusogen. Our lab determined the structures of gB and gH/gL. For gB, only the structure of the postfusion form of the ectodomain is known.