The Ralph Isberg Lab
Analysis of Bacterial Uptake & Growth within Mammalian Tissues
Traditionally, the work in our laboratory is directed toward investigating the interaction of microbial pathogens with host immune cells, as well as understanding how the host innate immune response deals with pathogen attack. In recent years we have begun to investigate drug resistance in nosocomial pathogens emphasizing a systems biology and network analysis approach. Much of the work of our laboratory is devoted to investigating intravacuolar growth of Legionella pneumophila within macrophages. We have also made recent inroads into understanding how the host responds to this organism. In addition, we are studying microbial colony formation within deep tissue sites, focusing on Yersinia pseudotuberculosis. Finally, we have focused on the evolution of antibiotic resistance of Acinetobacter baumannii, in hopes of developing novel therapies that overcome emerging drug resistance. The most exciting recent developments are the identification of a strategy for targeting tubular endoplasmic reticulum by L. pneumophila proteins and the development of a bioengineered tissue-free system that allows in vitro construction of abscesses harboring Y. pseudotuberculosis.
Intracellular growth of Legionella pneumophila
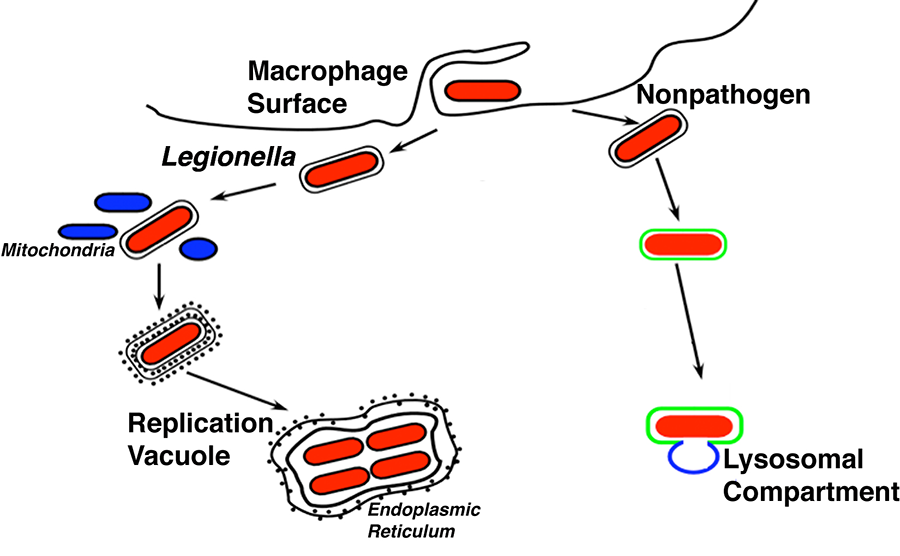
L. pneumophila is a pathogen adapted to grow within amoebae that also causes a severe pneumonia in humans known as Legionnaire’s disease. The bacterium avoids phagocyte killing by growing within a replication vacuole that initially bypasses the lysosomal network (Fig. 1). The bacterium is first internalized into a membrane-bound replication vacuole, with the initial event being recruitment of tubular endoplasmic reticulum. The replication vacuole eventually matures and the tubular structures are replaced by sheets of rough endoplasmic reticulum. In addition, host proteins that are involved in moving membrane from the endoplasmic reticulum to other compartments in the secretory apparatus are found associated with the replication vacuole.
Figure 1. This schematic shows the path of Legionella inside a cell as it replicates in membrane-bound compartment surrounded by rough endoplasmic reticulum. This is in contrast to a nonpathogen which is directed to the lysosome.
Formation of this replication vacuole requires 26 Dot/Icm bacterial proteins, which form an apparatus that translocates proteins into target host cells. Over the years we have devoted considerable work to identifying proteins that are translocated via this apparatus, which now number over 300. These translocated substrates appear to associate with cytoplasmic surface of the membranous compartment surrounding the bacteria. Our laboratory is attempting to construct a unified model for how these different bacterial proteins interact with host signaling pathways. Many of the proteins identified interact with host factors that control vesicular traffic from the endoplasmic reticulum to the Golgi, consistent with some steps in the replication vacuole biogenesis involving hijacking of vesicular material exiting from the endoplasmic reticulum.
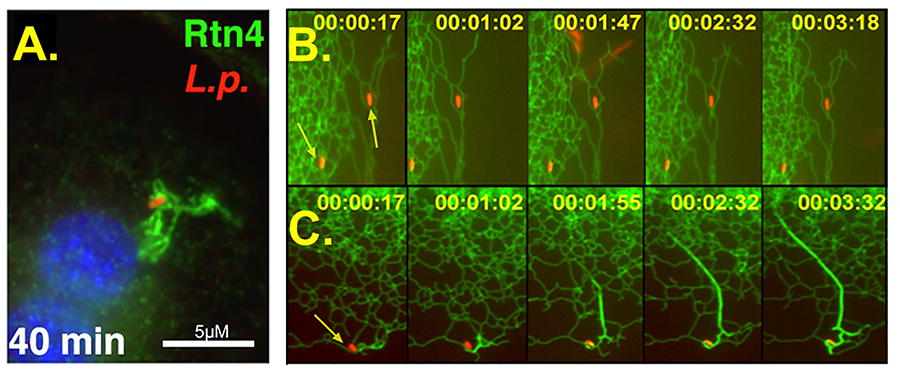
One important point of focus of the lab is on the control of host tubular endoplasmic reticulum function by L. pneumophila. Shortly after contact with host cells, the bacterium introduces the Sde family of proteins into host cells. These proteins encode a pathway that ubiquinates targets in host cells. The most dramatic consequence of the action of these proteins is to target the tubular endoplasmic reticulum protein Rtn4 (Fig. 2). After targeting of Rtn4 by the Sde proteins, Rtn4 rearranges to form a network of tubular endoplasmic reticulum (Fig. 2A, green) about the replication compartment (Fig. 2A, red). In the absence of Sde proteins, this does not occur (Fig. 2B). The response of cells to Sde introduction is rapid, with alterations in Rtn4 being witnessed within 2 minutes of contact with the bacterium (Fig. 2C).
Figure 2. These immunofluorescence pictures show Legionella (in red) within a replication vacuole associating with rearranged Rtn4 (green). Fig. 2A shows the bacterium 40 min after contacting the host cell. Figs. 2A and 2B are panels of real time video microscopy showing Rtn4 (green) and Legionella. In 2B, a mutant bacterium lacking Sde is unable to cause thickening and rearranging of Rtn4 tubules. In 2C, the wild type bacterium shows evidence of altering host cells within a minute. By 3.5 minutes, there is dramatic restructuring of Rtn4 about the bacterium.
Yersinia pseudotuberculosis control of host cell responses
Yersinia pseudotuberculosis is internalized by M cells overlying the intestinal Peyer's patches. Shortly after this event, bacteria are found exclusively extracellularly due to the production of Yops, bacterial proteins that are translocated via a secretion system needle (called a type III secretion system) that antagonize phagocytosis. Our work demonstrated that the primary bacterial-encoded factor that allows uptake both into M cells and into cultured cells is the outer membrane protein invasin, which binds multiple β1 integrin receptors. Our recent work has demonstrated that a key signaling molecule downstream from the integrin is the small GTPase, Rac1, which directs actin rearrangements in response to bacterial binding.
Recently, we have turned our focus away from these issues and have been investigating two critical aspects of Yersinia biology. The first involves how the host cell supports the function of the bacterial type III secretion system to allow translocation of the Yops via the needle inserted into the target cell. We conducted an RNAi screen to identify host proteins necessary to allow the needle to insert in host cells, which identified chemokine receptors and surface proteins linked via glycophosphatidylinositol tails. We are currently testing a model that these proteins send important signals to allow organization of a bacterial translocation pore in the host cell.
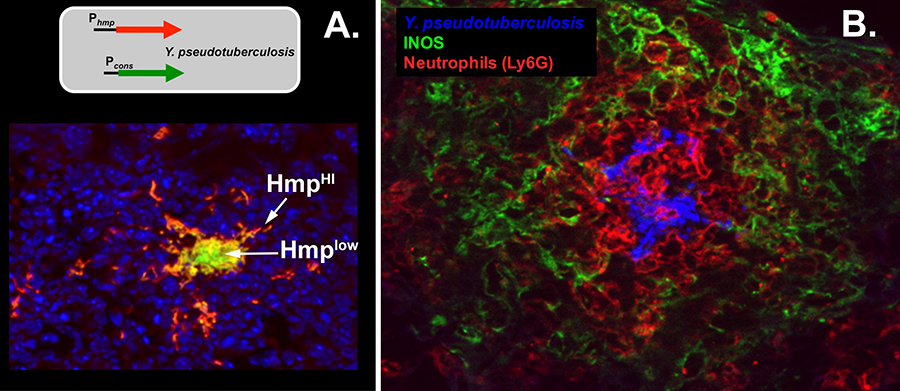
The second aspect of Yersinia biology that we study is the organization of bacteria and immune cells within tissues. The most important result that we have obtained is that the bacteria participate in social behavior within tissues while they grow as microcolonies in organs such as the liver and spleen. The best example of this is demonstrated by the expression of a protein called Hmp that detoxifies nitric oxide (NO) elicited by immune cells to eliminate bacteria. Yersinia growing around the edges of microcolonies express luxurious amounts of Hmp (seen as red, Fig. 3A) to detoxify NO. This provides shelter for bacteria in the center, who are protected from NO and as a result do not produce the HMP protein (seen as green or red, Fig. 3A). Surprisingly, the host cells that produce NO are not in direct contact with the bacteria, but form a layer separated from the bacteria (green, Fig. 3B). Therefore, growth of bacteria in tissues forms a structure akin to Dante’s inferno, with a series of spheres extending out from the growing bacteria (Fig. 3B). Our current goal is to totally reconstruct this system in an engineered microcolony system in the absence of host tissues, and analyze each of these populations at the molecular level.
Figure 3. Dante’s inferno: rings of Yersinia gene expression (panel A) surrounded by rings of immune cells (panel B). Panel A: A Y. pseudotuberculosis was constructed that harbors two reporters. The green reporter is constitutively expressed, the red reporter is expressed in the presence of NO. Mice were infected with this strain, and shown is a bacterial microcolony that forms in the spleen. The blue stain shows spleen cells. The red identifies bacteria that are exposed to NO. Green or yellow are bacteria that experience little or no NO. Panel B: stained are Y. pseudotuberculosis (blue), neurophils (red) and NO-producing cells (green).
Determinants of multi-drug resistance in Acinetobacter baumannii
The emergence of highly antibiotic-resistant infections represents one of our greatest threats to public health. Many multidrug-resistant (MDR) pathogens cause diseases that are primarily associated with hospitalization. These infections are particularly problematic in immunocompromised patients such as those undergoing chemotherapy for cancer or transplantation, for which successful treatment of opportunistic infections is critical. One of the most dangerous emerging MDR pathogens is A. baumannii. This Gram-negative bacterium is the cause of a number of hospital-acquired diseases including septicemia resulting from indwelling devices, pneumonia, and wound infections. A. baumannii infections are especially troublesome in intensive care units (ICUs), as the pathogen is now among the 5 most common causes of ventilator-associated pneumonia in US hospitals, with high associated mortality. Over recent years an alarming increase in drug resistance rates has been observed in this organism.
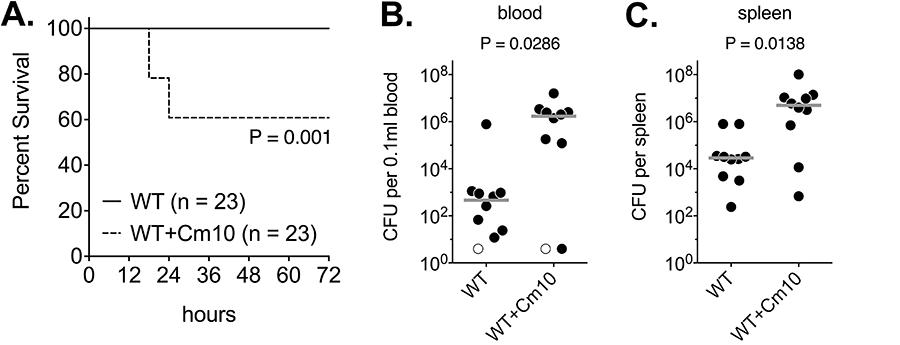
Our laboratory has been focusing on several aspects of A. baumannii drug resistance and virulence in animals. One project has been the identification of a regulatory locus called BfmRS, which demonstrates that the potential to cause disease and the ability to acquire antibiotic resistance are linked and controlled by the same regulatory pathway. BfmRS, which is totally dispensable for the bacterium to grow outside of hosts, regulates bacterial envelope biogenesis. Its absence causes the bacterium to be highly sensitive to attack by the host innate immune system, as well has hyper-sensitive to antibiotics. This project also demonstrated that inappropriate antibiotic treatment, such as exposing bacteria to sublethal amounts of chloramphenicol, increases the virulence of the organism, emphasizing the importance of proper dosing during the treatment of disease (Fig. 4).
Figure 4. Low level antibiotic treatment stimulates virulence-associated properties of A. baumannii. Panel A: Treatment of A. baumannii with chloramphenicol (Cm10) for 1 hr converts a low-virulence bacterium into a pathogen. Panels B and C: Bacteria treated for one hour with chloramphenicol show accumulation in the blood and spleens in infected animals.