The Linden Hu Lab
Below are some of the ongoing bench research projects in the laboratory. In addition to the projects listed below, the laboratory is working on clinical/translational projects to study xenodiagnoses of Lyme disease after treatment with antibiotics and to develop new strategies for development of a human vaccine for Lyme disease.
Borrelia burgdorferi
Escape from Innate and Adaptive Immunity by B. burgdorferi
Lyme disease is the most common vector borne disease in the U.S. and continues to expand in both inci-dence and geographic distribution. Although tremendous strides have been made in the understanding of Lyme disease and of B. burgdorferi pathogenesis, until recently researchers interested in studying the role of specific genes have been limited to painstaking efforts to mutate genes and examine the mu-tants individually for infectivity phenotype. Microarrays and proteome analyses of B. burgdorferi have revealed there are significant changes in gene expression in response to shifts in environmental conditions. However, to date, only a minority of these factors have been studied in vivo due to the difficulty in performing these studies. High throughput strategies that accelerate identification of virulence determinants, essential genes and adaptive strategies of other bacteria have only recently become available for use with B. burgdorferi. We are taking advantage of advances in B. burgdorferi genetics and massively parallel sequencing to perform genome-wide screening to identify virulence determinants contributing to growth within a mammalian host. We use transposon mutant libraries in B. burgdorferi to quantitatively identify relative fitness of each mutant during infection—essentially combining individual competitive fitness experiments into much larger scale experiments. This provides a major advantage compared to traditional gene inactivation studies in B. burgdorferi which can only report results as either having an “infective” or “non-infective” phenotype without more subtle examinations of partial fitness. To date, we have performed in vivo screens of the transposon libraries in mice with defects in innate (MyD88 deficient mice) and adaptive (severe combined immunodeficiency or SCID mice) to determine whether these defects in host immunity “rescue” transposon mutants with fitness defects. We are currently working on identifying the mechanism by which the identified genes contribute to immune escape.
We have also performed focused studies on the role of specific genes in evading killing by reactive oxygen species (ROS) and reactive nitrogen species (RNS). ROS/RNS are important components of innate immune defenses in all of the different B. burgdorferi reservoir hosts. NADPH phagocyte oxidase is the main generator of ROS and inducible nitric oxide synthase (iNOS) is the main generator of RNS in mammals. Arthropod ROS is generated by dual oxidase and NADPH oxidases and have been shown to play a role not only in immune defense against microbes, but also in development. Arthropod RNS are generated by activity of nitric oxide synthase and nitrophorins, which are nitric oxide containing heme molecules. RNS may also be released at the tick bite site and contribute to vasodilation that assists with tick feeding. We have now identified subsets of B. burgdorferi genes important in survival against ROS and RNS. We have confirmed that many of these genes are important in survival of the organism during the mammalian and/or tick phase of its lifecycle. Additionally, we have used the transposon library to test gene fitness in the tick host and found that a large percentage of the genes involved in ROS/RNS defenses are critical for survival in the tick. These studies have yielded a wealth of targets that will advance our understanding of how the bacteria has adapted to different immune defenses of its hosts and the strategies it uses to recognize and evade. The laboratory is collaborating with Drs. Jon Skare and Jennifer Hyde at Texas A & M to study the role of these proteins in evading ROS/RNS killing in the different hosts.
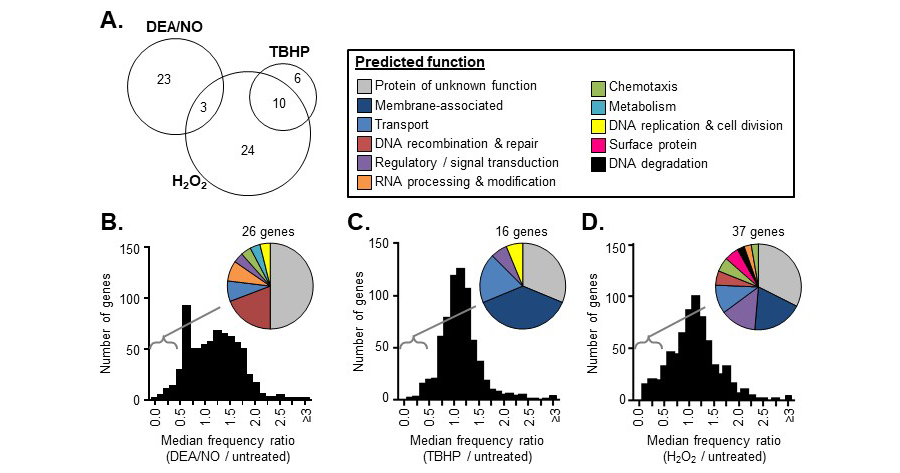
Figure 1. Overview of in vitro Tn-seq screen results. A frequency ratio (frequency in the treated library / frequency in the untreated library) was determined for each Tn mutant following DEA/NO, TBHP, or H2O2 exposure. An overall frequency ratio was also determined at the gene level by aggregating all of the sequence reads mapping within in the same gene. (A) The number of genes with overall frequency ratios <0.5 in both replicates of the DEA/NO, TBHP, or H2O2 Tn-seq screens. (B-D) The distribution of overall frequency ratios for each gene after DEA/NO (B), TBHP (C), or H2O2 (D) exposure. Pie charts indicate the predicted functions of the genes with overall frequency ratios <0.5 in each case.
Role of host tolerance in bacterial persistence
While the ability of certain pathogens to invade hosts and cause long term systemic infections is typically ascribed to attributes of the pathogen in evading host immunity, hosts defenses may also choose to declare “détente” and cease to attack the invader if the cost of continued attack is greater than the cost of co-existence (e.g. when continued activation of the host response causes more damage than the organism). B. burgdorferi, the causative agent of Lyme disease, is able to establish prolonged infections in its mammalian hosts. It is not known to produce harmful toxins or enzymes and most of the damage in response to human B. burgdorferi infection is thought to be due to the effects of the immune response. Peromyscus mice, the main natural host for B. burgdorferi, once infected, remain infected for life, yet display no symptoms from infection and little to no evidence of an inflammatory response to the presence of the bacteria. Even inbred mice that develop strong inflammatory responses to B. burgdorferi with initial evidence of carditis and arthritis, resolve the inflammation over time despite the continued presence of live bacteria. While part of the reduced inflammation is due to a reduction in the number of organisms through killing by the adaptive immune system, there is evidence to suggest an important component is a change in the responsiveness of host innate immunity with pro-longed exposure to the organism.
The innate immune system plays an important role in mediating inflammatory responses to B. burgdorferi. B. burgdorferi components are recognized by multiple toll-like receptors (TLRs) and cytosolic receptors including TLRs 2, 5, 7, 8, 9 and Nod2. In vitro, loss of TLR or Nod2 signaling affects downstream release of pro-inflammatory cytokines in response to exposure to B. burgdorferi. However, interestingly, animals deficient in these innate immune receptors or adaptor molecules that transduce signals from TLRs-- including but not limited to TLR2, MyD88, CD14, TRIF-- do not show reductions in inflammation (e.g. arthritis) during infection with B. burgdorferi and often show increased inflammation and tissue levels of inflammatory cytokines. In many cases, bacterial loads are similar between wild type and knockout animals suggesting that the increased inflammation is not the result of reduced control of infection. This suggests that the major role for these innate immune molecules is in dampening rather than augmenting inflammation. One major difference between in vitro studies and the in vivo infections is that the in vitro experiments are typically conducted by measuring responses minutes to hours after exposure to the organism. In the animal, inflammation is typically assessed 2-6 weeks after infection. We have evidence that more prolonged exposures in vitro result in the development of innate immune “tolerance” to stimulation by B. burgdorferi. Cells pre-exposed to B. burgdorferi or other innate immune stimuli, become less responsive upon re-exposure, releasing less pro-inflammatory cytokines and more anti-inflammatory cytokines such as IL-10. Using techniques such as micro-RNA seq, we are currently working to understand the mechanisms by which host cells moderate tolerance to B. burgdorferi and avoid self-induced injury during long term infection.
Mechanisms for intracellular regulation of TLR2 signaling
The innate immune response protects the host from microbial invaders through recognition of molecular patterns specific to pathogens or to the damage they create. Among the best studied of these pattern recognition receptors are the TLRs. TLR2 heterodimerizes with either TLR1 or TLR6 to recognize components of a diverse array of pathogenic bacteria including Staphylococcus aureus, Streptococcus pyogenes, Mycobacterium tuberculosis and B. burgdorferi. The TLR2 mediated responses to these organisms vary substantially. How TLRs are capable of generating specific responses to individual bacteria recognized by the same receptor is a major focus of the field. Although the repertoire of available signaling molecules downstream of TLRs is rather limited in number, they assemble to generate a surprising amount of diversity. While early models of TLR2 interactions with its ligands were based on the concept of a single ligand reacting with a single receptor at the plasma membrane surface, it is now recognized that multiple adaptor molecules and co-receptors can interact with TLR2 or its ligands, altering specificity of both recognition and response. One mechanism used by cells to control responses is to through limiting assembly of signaling platforms to specific cellular compartments.
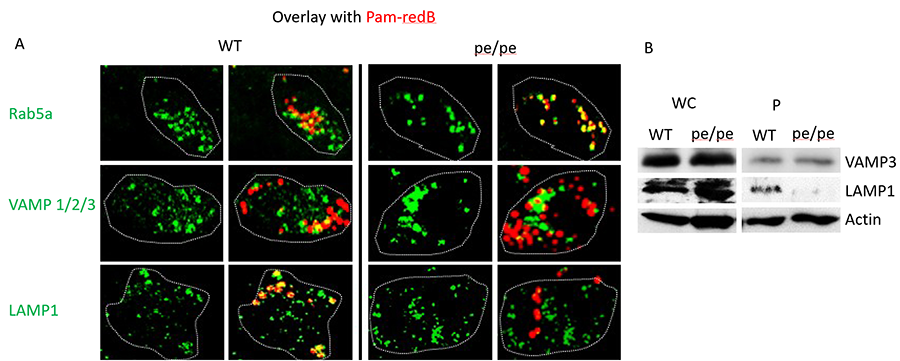
Figure 2. AP-3 traffics TLR2 ligands from early to late phagosomes. (A) BMDM from wild-type (WT) and AP-3–deficient mice (pe/pe) were stimulated with Pam3CSK4-biotin–coated red fluorescent streptavidin beads (Pam-redB) for 60 min. Cells were fixed in 1% paraformaldehyde and stained in 2% goat serum and 1% saponin. Primary Abs were used as indicated, followed by secondary Ab conjugated to Alexa 488 (green). Cells were mounted on cover slips, visualized by confocal miscroscopy using a ×63 oil objective, and analyzed by Fiji (ImageJ). Dotted lines indicate cell borders. Shown are representative images of three independent experiments. Images were taken at original magnification ×4. (B) BMDM from WT or AP-3–deficient pe/pe mice were stimulated with magnetic Pam-B for 60 min and phagosomes isolated by magnetic pull down. Whole-cell (WC) input and phagosomal fractions (P) were run on SDS-PAGE and blotted with Abs against VAMP-3, LAMP-1, and actin. Shown is a representative blot of three independent experiments.
While individually, much is known about both intracellular trafficking and innate immune signaling, we are just beginning to understand the intersections between these two disciplines. Localization to early endosomes is an important determining step in the fate of internalized cargo and an important modulator of signaling. we have shown that changes in intracellular location result in alterations in innate immune outputs initiated by TLR2. we showed by altering intracellular trafficking through deletion of an important trafficking molecule, AP3, that localization to a LAMP1+ lysosome is necessary for the TLR2 mediated activation of some cytokines (e.g. IL-6) but not others (e.g. TNF-α) 5. We also showed that the localization of adaptor molecules to a compartment is not sufficient to initiate TLR2 mediated IL-6 signaling suggesting there are additional event(s) required to initiate TLR2 dependent signaling from the phagosome. To identify potential events involved in activating signaling from these LAMP1 compartments, we performed an unbiased screen of vesicular trafficking molecules using a siRNA library. Using this system, we have uncovered multiple novel targets that are involved in activating signaling downstream of TLR2 from LAMP1 compartments. Current work in the laboratory involves delineating the role of each of these components and how they interact to activate signaling in response to TLR2 ligands.
Reduction of Vector and Reservoir Competence for Carriage of B. burgdorferi
The incidence and geographic distribution of Lyme disease in the US has increased steadily since its first description in 1977. Efforts to stem the spread of the disease through controlling the population of its tick vector and/or the mouse reservoirs of the disease have met with only limited success. The only approved human vaccine to protect against Lyme disease was removed from the market by its manufacturer further highlighting the need for new approaches to controlling the disease. The incidence and geographic distribution of Lyme disease in the US has increased steadily since its first description in 1977. Efforts to stem the spread of the disease through controlling the population of its tick vector and/or the mouse reservoirs of the disease have met with only limited success. The only approved human vaccine to protect against Lyme disease was recently removed from the market by its manufacturer further highlighting the need for new approaches to controlling the disease. In this project, we are developing an orally-available vaccine targeted towards the mouse and tick reservoirs of the disease. Prior attempts to vaccinate wild animals have been hampered by the lack of an efficient, oral delivery system which is both stable under natural environmental conditions and can generate an intense immune response. The photo (courtesy of our collaborator Dr. Sam Telford, Cummings School of Veterinary Medicine) illustrates a mouse next box for distribution of baits. We are studying the use of a variety of viral vectors as a delivery mechanism for a targeted mouse vaccine. In addition, we are working to develop new vaccines that combine “traditional” approaches that target the pathogen with novel approaches that target the tick vectors. Preliminary data has shown that uptake of antibodies directed at tick proteins by ticks during a blood meal can inhibit feeding, prevent transmission of pathogens and even result in tick mortality. A tick directed vaccine may be particularly relevant for tick-transmitted pathogens such as Babesia microti and Anaplasma phagocytophilum where there are no advanced vaccine targets.
Figure 3. Mouse nest box.
In conjunction with Kim Lewis’ laboratory at Northeastern, we are also working on the development of new, non-human antibiotics that may be targeted for use as reservoir clearing compounds. Use of antibiotics such as doxycycline to clear infections from ticks and mice has been shown to be highly effective. However, concerns for the development of resistance in both B. burgdorferi and other tick-borne diseases such as A. phagocytophilum have limited the use of this approach. The use of compounds that are not used for treatment of human infections and that do not engender cross-resistance with currently used human antibiotics is a new approach that may lead to successful eradication of the organism.
Porphyromonas gingivalis
Periodontitis is an inflammatory condition that, if untreated, results in reduction of gingival tissue, destruction of the periodontal ligament, alveolar bone resorption, and loss of teeth. Porphyromonas gingivalis, an important pathogen in the development of periodontitis, interacts bi-directionally with other members of the oral microbial community. The organism modulates both the host immune system and the composition of the oral microflora resulting in a dysbiosis that leads to inflammation and bone loss. Counteracting these effects, certain “commensal” bacteria in the oral flora help maintain oral health by affecting the ability of P. gingivalis to colonize dental tissues.
A growing body of literature indicates that bacteria in the oral microbiome communicate with each other affecting behavior within the bacterial community. It has been clearly shown that species such as streptococci and Fusobacterium nucleatum, that may be minimally pathogenic by themselves, act as scaffolds in the formation of the subgingival biofilm by providing attachment for pathogens such as P. gingivalis, thus enhancing their virulence. Much less is known about how bacteria in the oral flora may inhibit the virulence of pathogenic species. Recent advances in technology, such as next generation sequencing, now allow analysis of shifts in the oral microbiota providing a broad view of the microbiome associated with health and disease. However, microbiome studies do not provide insight into the molecular interactions between different species and strains that result in these shifts.
We have found that strains of bacteria from the oral flora as well as lactic acid bacteria (LAB), a class of bacteria commonly used as probiotic agents, are capable of suppressing virulence of the dental pathogen P. gingivalis by a variety of mechanisms. We have found that these effects are strain dependent and not just species dependent. Utilizing chemical mutagenesis, transposon mutagenesis and whole genome sequencing technologies, the laboratory is currently working to identify the mechanisms by which specific strains of bacteria are capable of inhibiting the virulence of P. gingivalis and to understand which genes are essential for P. gingivalis survival and virulence under different conditions.
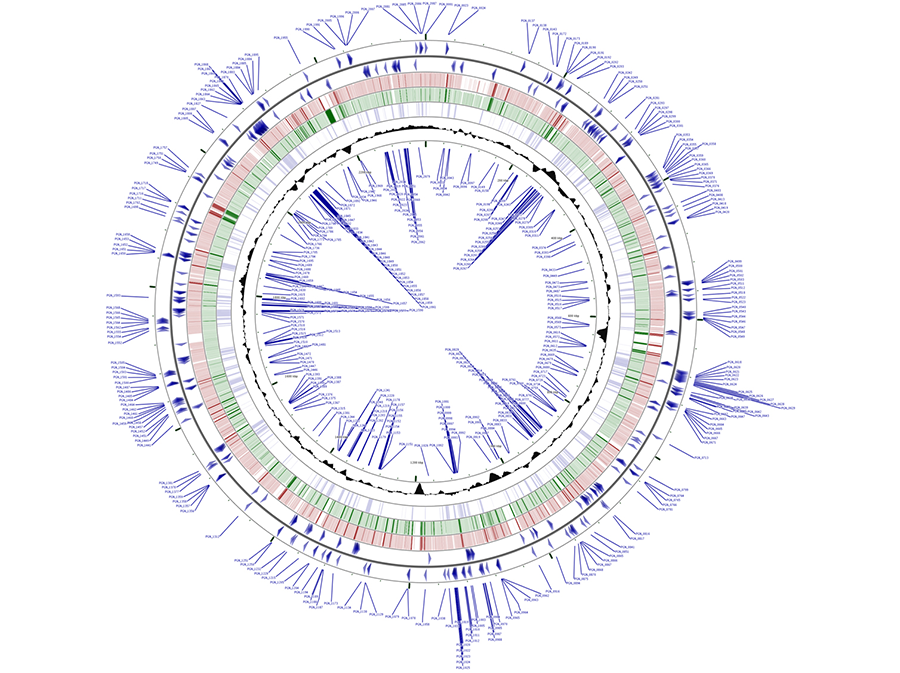
Figure 4. Mapping of the P. gingivalis essential genes. In blue in the outermost and innermost rings are shown the putative P. gingivalis essential genes identified by transposon mutagenesis of strain ATCC 33277. Blue arrows depict the orientation of the essential genes. Genetic loci for positive strand (outer set of arrows) are shown in the outermost circle and genetic loci for the negative strand are shown in the innermost ring. In red are coding sequences for P. gingivalis strain W83 and in green are coding sequences for strain TDC60. Shaded black area represents GC-content for given regions. CGViewer was utilized to visualize the entire circular genome of P. gingivalis strain ATCC 33277 with the putative essential genes labeled. NCBI FASTA files of P. gingivalis strains W83 and TDC60 were used for BLAST matching to the ATCC 33277 base genome.