The Shumin Tan Lab
Environmental Cues in Mycobacterium tuberculosis-Host Interactions
Mycobacterium tuberculosis (Mtb) is an extremely successful pathogen that remains a key public health problem, with broad impact on population and economic development. We seek to understand the molecular mechanisms of Mtb-host interactions, with a focus on the host environmental signals that serve as cues for Mtb during host colonization. Knowledge of how environmental cues in vivo are integrated with core aspects of Mtb biology, such as its metabolism and growth, is fundamental to our understanding of Mtb infection and tuberculosis disease progression.
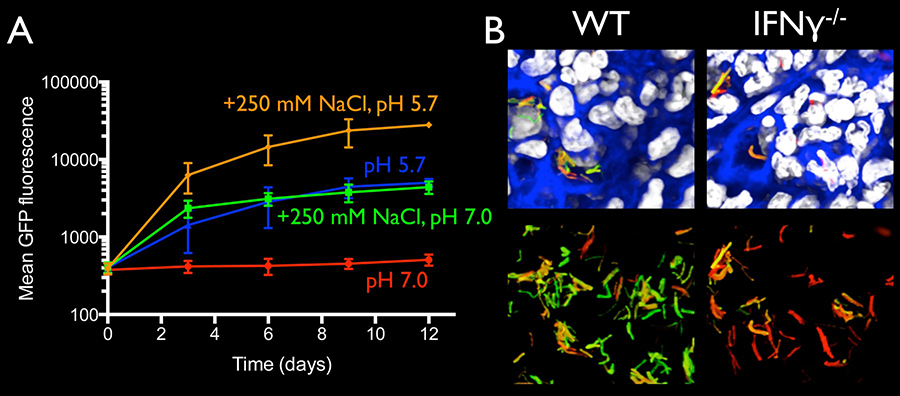
One area of particular interest with environmental cues is the understudied concept that local concentrations of abundant ions can profoundly impact a pathogen’s ability to colonize its host. This concept stems from our earlier discovery that chloride concentration is inversely related to pH during phagosomal maturation, and that Mtb responds synergistically to these two cues, with the response reflecting the host’s immune status (Figure 1). We have also found that Mtb responds to potassium, and are continuing to build on these findings, exploring how Mtb response to these disparate signals are linked, and the intrinsic relationship of these environmental responses to bacterial metabolism and growth. Elucidation of nodes critical to the bacterium’s ability to coordinate its response to multiple signals open up potential avenues for targeting Mtb to shift the balance of infection in favor of the host.
Figure 1. (A) Mtb responds synergistically to chloride and pH, as illustrated by differential induction of GFP signal in reporter Mtb (rv2390c’::GFP) engineered to express GFP in response to increased chloride concentration or decreased pH in its environment. (B) In a murine infection model, this reporter signal (green) is higher in the bacteria present in lungs of wild type (WT) versus immune-deficient interferon gamma (IFNγ) knockout hosts 4 weeks post-infection. All Mtb were also engineered to constitutively express mCherry (smyc’::mCherry, red) for visualization of the bacteria in the lung tissue. Host nuclei are in grayscale, and phalloidin staining of f-actin is shown in blue in these 3D confocal images. (Adapted from Tan et al, PLoS Pathog 9: e1003282, 2013)
Mycobacterium tuberculosis Infection Heterogeneity
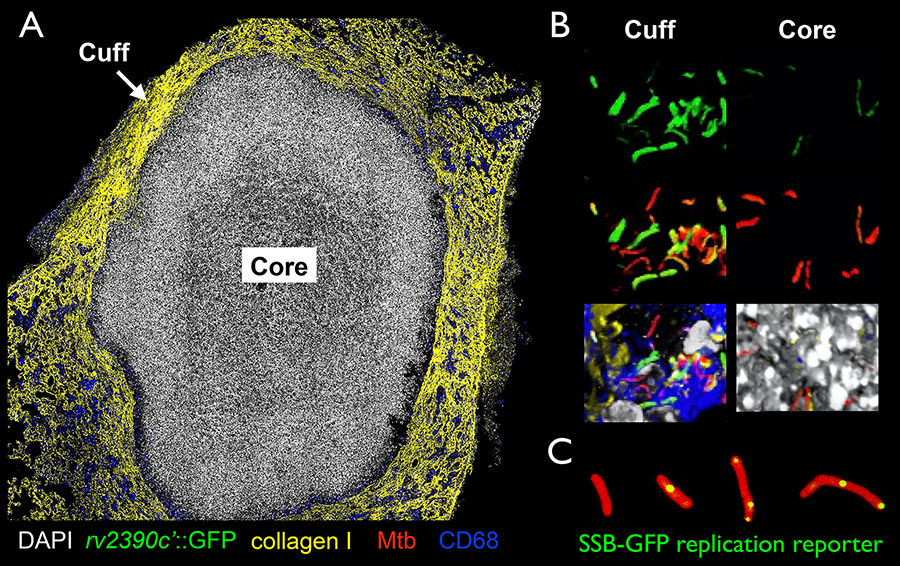
A hallmark of Mtb infection is the marked heterogeneity present across multiple facets. Our in vivo studies utilizing reporter Mtb strains have served to illustrate the extent of this heterogeneity – for example, there is non-uniformity in environmental signals even within sublocations of a single structured lesion (Figures 2A and 2B). We are pursuing studies to understand how heterogeneity in environmental cues impact on Mtb growth and disease progression and treatment outcome, utilizing unique reporters such as a replication reporter Mtb strain (SSB-GFP) that marks bacteria undergoing active DNA replication with green foci (Figure 2C). Reporter Mtb strains allow us to examine questions of heterogeneity at the individual bacterium level, while maintaining spatial context and the full complexity of in vivo host infection. By exploiting these strains in infection models and combining it with thick tissue imaging and 3D confocal reconstruction, we aim to elucidate how environmental signals may drive Mtb infection heterogeneity, and factors that distinguish an infection focus as conducive or restrictive for Mtb growth in vivo.
Figure 2. (A and B) Confocal images of a caseous necrotic lesion from an 8-week murine infection, illustrating the intra-lesion environmental heterogeneity observed during Mtb host infection. (A) shows an overview image of the lesion, while (B) shows 3D confocal images from the lesion cuff and core. Reporter signal is in green (rv2390c’::GFP), all Mtb are in red (smyc’::mCherry), nuclei are in grayscale (DAPI), collagen I staining is in yellow, and macrophages (CD68) are in blue. (C) SSB-GFP replication reporter marks Mtb undergoing active DNA replication with green foci. The bacteria also constitutively express mCherry via a smyc’::mCherry construct (red).