The Caroline Genco Lab
Immune Mediated Diseases and the Microbiome
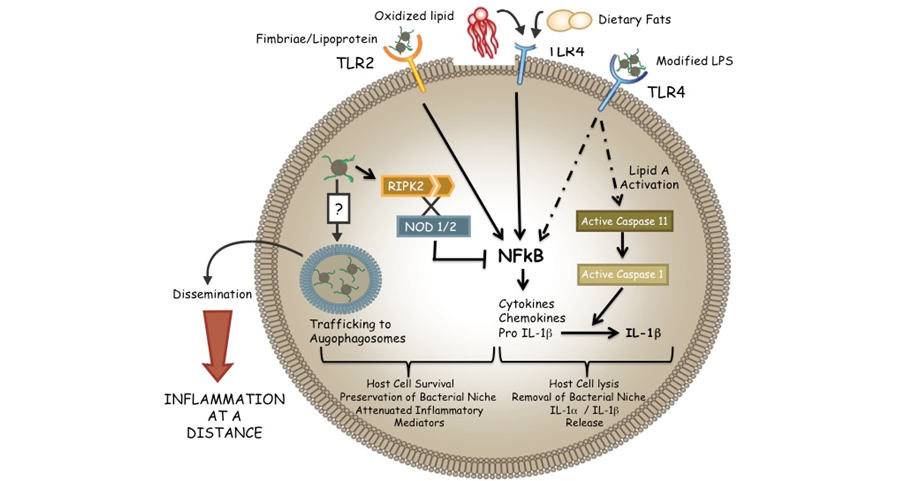
A primary focus of the laboratory is in the area of chronic inflammation and the role of the microbiome in systemic inflammatory disorders. Current studies are centered on the role of the innate immune system in chronic inflammation and on the association between microbially induced inflammation and atherosclerosis with new studies in the area of pancreatic cancer. We have defined the role of specific innate immune signaling pathways in immune cells that contribute collectively to pathogen-induced chronic inflammation. We are examining in vitro model systems for endothelial cells, macrophages / monocytes, and dendritic cells. Using defined animal models of inflammation we are characterizing the roles of innate immune pathways in inflammatory processes in vivo. The long-term goals of this work is to define the interplay between the innate and adaptive immune system and the regulatory pathways that contribute to endogenous and microbial ligand induced chronic inflammation.
Figure 1. Model of Porphyromonas gingivalis infection.
Innate Immune Responses to Mucosal Pathogens
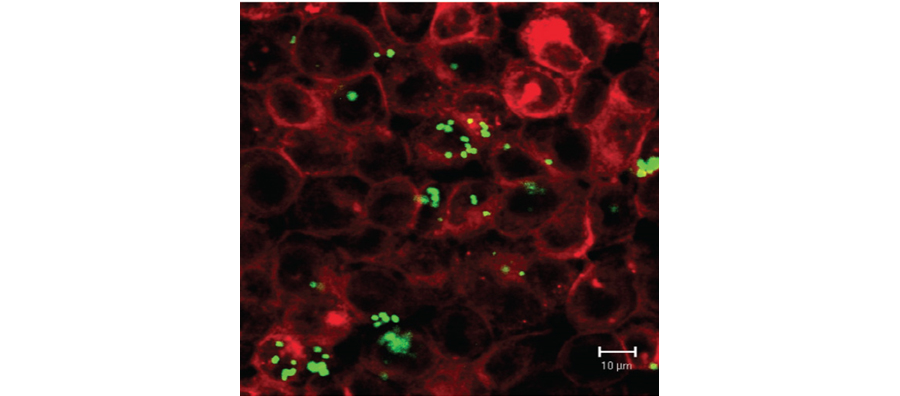
We have examined the interactions of several mucosal pathogens with both phagocytic and non-phagocytic cells and have defined the role of specific TLRs in recognition of both purified microbial ligands and defined bacterial mutants. We have defined signaling pathways in human endotheilal and epithelial cells and macrophages/monocytes. We have also examined immune signaling pathways in mouse and human platelets. Through the construction of bacterial mutants and using defined microbial ligands we have defined unique mechanisms by which pathogens evade host immunity. Much of this work has utilized human tissue models systems and more recently mouse models of colonization and inflammation to validate our in vitro finding. New work is aimed at defining markers of dendritic cell mediated immune homeostasis and atherosclerosis in several existing human observational cohorts. Our work examining inflammatory responses to bacterial pathogens also extends into human correlates in existing cohorts. This work has focused on gonococcal pathogenesis in cohorts of subjects both in the US and in Nanjing, China.
Figure 2. Confocal imaging of Neisseria gonorrhoeae infection of endocervical cell line E6/E7
Regulatory Mechanisms in Bacterial Pathogens
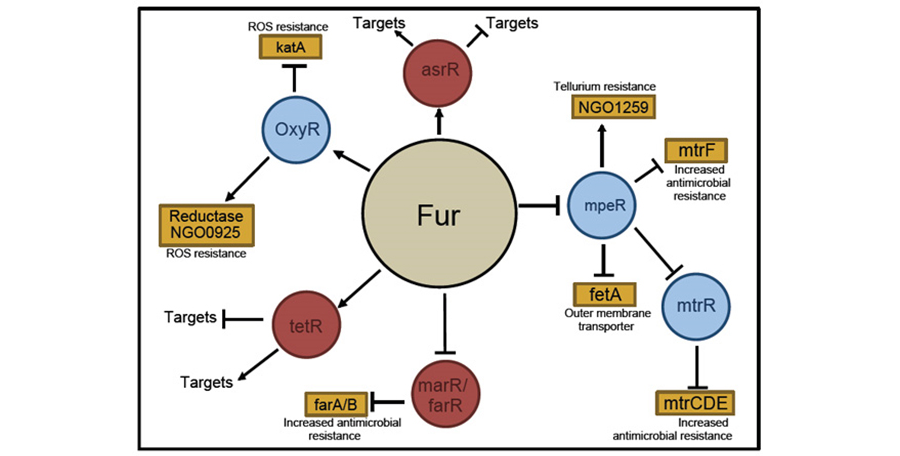
Work in the area of regulatory mechanisms in bacterial pathogens is focused on understanding mechanisms utilized for bacterial colonization, and in particular in the ability of in vivo environmental factors to modulate bacterial gene expression. Transcriptional regulatory mechanisms have been defined on a global level in the pathogenic Neisseria species. We have established that the expression of virulence factors in these organisms is controlled by a global regulatory protein (ferric uptake regulator protein, Fur). We established that the gonococcal Fur protein functions as a global regulatory protein to either repress or activate expression (directly or indirectly) of genes involved in pathogenic pathways. We identified several Fur-controlled genes that code for additional regulatory proteins, thus potentially expanding the Fur regulon via control of other DNA-binding proteins.
Figure 3. Fur regulation of N. gonorrhoeae gene expression