The Claire Moore Lab
Maturation of Eukaryotic mRNA
Our lab studies mRNA 3' end processing, an essential step in mRNA biogenesis in which primary transcripts are cleaved and then further processed by the addition of a tract of adenosine residues. The execution of this processing step prevents readthrough transcription from interfering with DNA elements such as promoters, centromeres, and replication origins. In addition, acquisition of the poly(A) tail is important for accumulation of mature mRNA, its export from the nucleus, its utilization in translation of protein, and its timely removal when the mRNA is no longer needed by the cell. Maturation of mRNA 3’ ends also serves as an important point at which the cell can regulate the type and amount of mRNA derived from a particular gene. Furthermore, it has been linked to other essential processes such as DNA repair and tissue-specific protein expression. Mistakes in polyadenylation can impact on all of these processes.
The goal of our research is to determine the molecular mechanisms of mRNA polyadenylation, its regulation in response to changes in cell status, and its coordination with other nuclear processes such as transcription and DNA damage repair. We use biochemistry, molecular biology, genetics and whole-genome expression analysis to study these issues. Our work has led to the identification and functional analysis of many components of the processing complex, the demonstration of its regulation by post-translational modifications, and the characterization of novel ways in which it interfaces with transcription, mRNA export and mRNA quality control. The research in the Moore lab pursues three inter-related themes as described below.
Molecular Mechanism of mRNA 3’ Polyadenylation
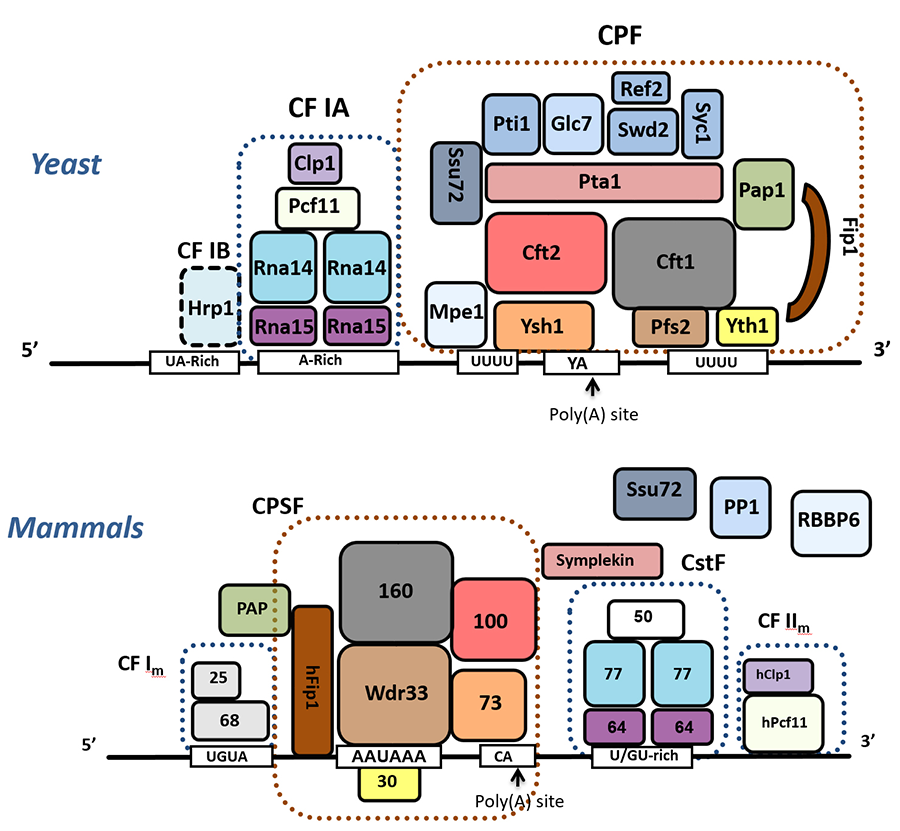
Our early research focused on identification of components of the cleavage/polyadenylation complex (Figure 1) and defining how different subunits contribute to functions such as enzymatic activity and recognition of signal sequences on the RNA that specify the poly(A) site. Our work also showed how some subunits provide scaffolding functions that help organize the multi-subunit factors, mediate cross-factor interactions to stabilize the complex at the poly(A) site, or facilitate transitions between different steps in a cycle of cleavage and polyadenylation. This detailed biochemical knowledge allowed us to appreciate the remarkable conservation of this process across eukaryotic species. It has also been important in helping us understand how the activity of the complex is influenced by post-translational modifications and by interaction with other cellular machineries such as the transcription elongation complex and mRNA export factors.
Figure 1. Composition of the cleavage/polyadenylation complexes from yeast and mammals. Conserved subunits are indicated in similar colors. In yeast, CPF contains the enzymatic activities - the endonuclease Ysh1 and the poly(A) polymerase Pap1. The CF IA and Hrp1 factors help position CPF at a poly(A) site.
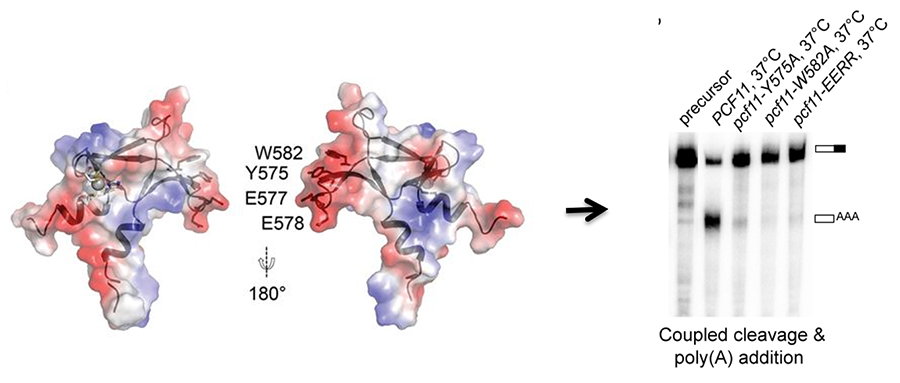
Collaborations with structural biologists help us understand how structure contributes to function of the mRNA 3' end processing complex. An especially rewarding synergy from these collaborations is that when the structures define critical residues, our mutagenesis and processing assays can then test function (Figure 2), and vice versa, the reason certain mutations perturb function becomes clear once the structure is available. We are now investigating how the processing factors are assembled from individual subunits.
Figure 2. The conserved C-terminus of Pcf11 forms a novel zinc-finger structure that is essential for mRNA 3’ end processing and links CF IA to CPF (Yang, et al, 2017)
Coupling mRNA 3' End Processing with Transcription
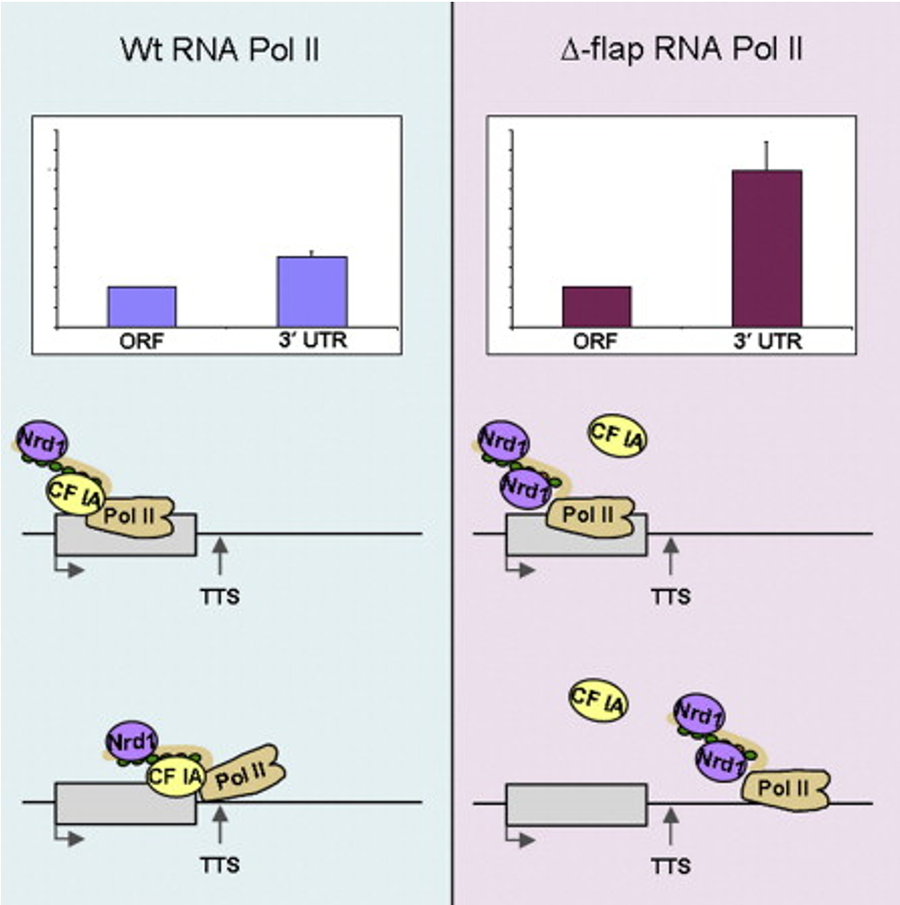
Much of our recent work focuses on how mRNA 3’ end processing is coordinated with transcription to keep mRNA synthesis balanced with the cell’s needs. For example, a major accomplishment was identification of the surprising dual nature of Ssu72. We showed that this subunit of CPF functions in transcription as a phosphatase to remove phosphates from the C-terminal domain of RNA Polymerase II (Pol II) that are important for elongation but would block re-initiation of transcription. In a role that does not require its phosphatase activity, Ssu72 blocks a domain of the Pta1 subunit of CPF from inhibiting processing. We also demonstrated that the conserved flap loop of Pol II, located near the RNA exit channel, mediates Pol II interaction with CF IA, thereby providing the first evidence that a structural feature of Pol II in addition to the C-terminal domain is important for termination factor binding and function (Figure 3).
Figure 3. The evolutionarily conserved Pol II flap loop contributes to proper transcription termination of short yeast genes by interacting with CF IA (Pearson & Moore, 2014).
Our latest work is defining how 3' end processing factors promote transcription elongation and transcription-coupled DNA damage repair, regulate the stress response in ways that do not involve their processing activity, and suppress expansions of trinucleotide repeats that can cause disease.
Regulation of polyadenylation - Post-translational Modifications
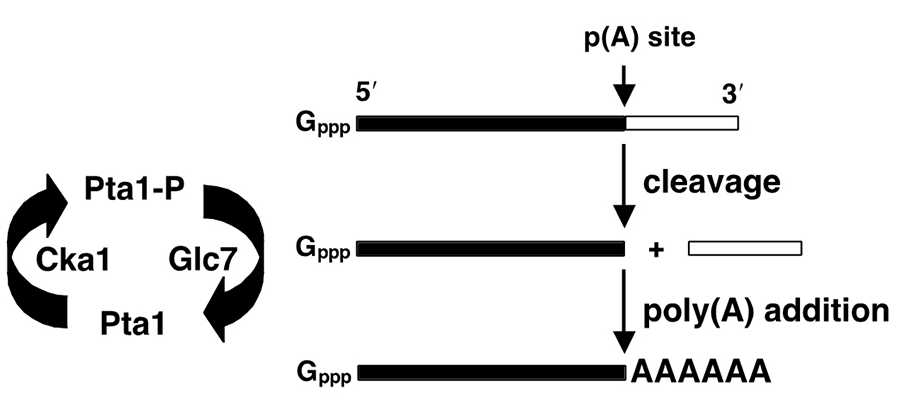
A major interest of our lab is how the activity of the processing complex is regulated through post-translational modifications. Our work has shown that the poly(A) addition step is inhibited by phosphorylation of the Pap1 and Pta1 subunits of CPF, and that this is counteracted by another subunit of CPF, the Glc7 phosphatase (Figure 4).
Figure 4. Phosphorylation regulates mRNA 3’ end processing (He & Moore, 2005).
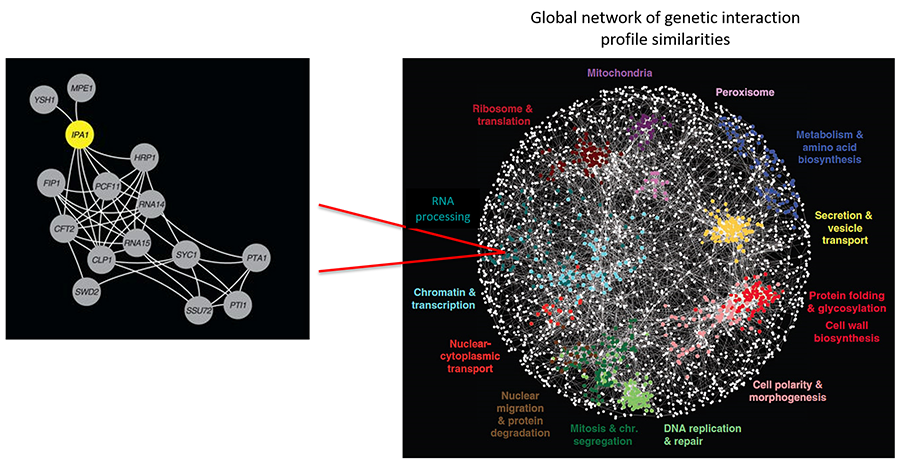
This cycle of addition and removal of phosphate could help the complex move from the cleavage to the poly(A) addition steps, and then release from the polyadenylated mRNA to begin another round of processing. Our ongoing work is aimed at understanding the role of ubiquitination in mRNA 3’ end processing. Application of synthetic genetic array analysis is allowing us to identify novel regulators of post-translational modifications such as Ipa1 (Figure 5). We are currently investigating how Ipa1 regulates the levels of the Ysh1 endonuclease, a protein at the heart of the processing complex.
Figure 5. Synthetic genetic array analysis identifies a new polyadenylation regulator (Costanzo, et al, 2016).
Regulation of polyadenylation - Alternative Polyadenylation
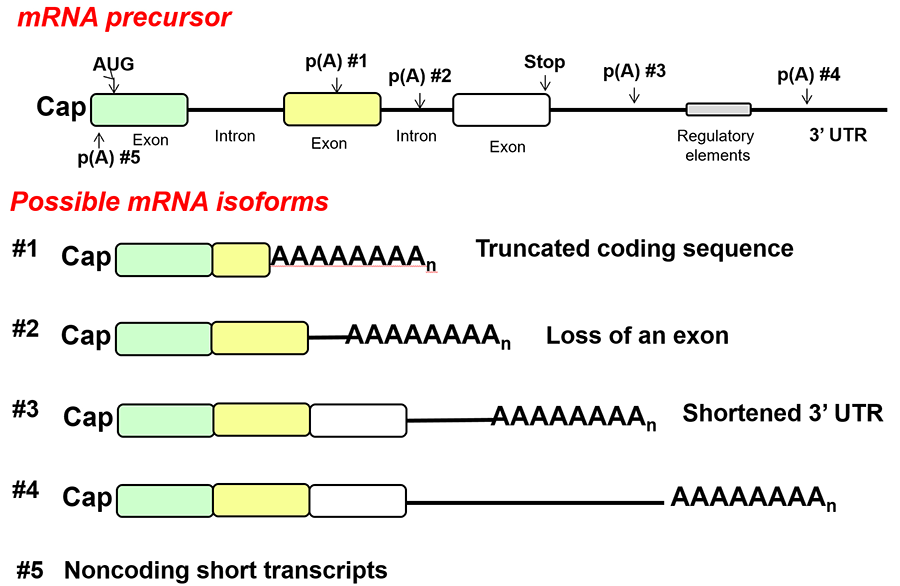
Many eukaryotic genes have multiple poly(A) sites, and the choice of poly(A) site can determine the stability, translatability, and cellular localization of the mRNA, as well as the amount of coding sequence available for translation (Figure 6). One of our goals is to understand the mechanism and consequences of alternative polyadenylation and how it contributes to the normal cell response to external signals as well as to human diseases such as cancer. A change in the profile of different mRNA isoforms could be due to the concentration of processing factors in the cell, regulatory proteins that direct the processing complex to specific sites, a change in transcription elongation between poly(A) sites, or changes in the relative stability of different length mRNAs. However, for most cases of alternative polyadenylation, the relevant mechanisms are not known. We are combining molecular biological approaches with computational analysis of whole-genome data sets to probe regulation of alternative polyadenylation in tumorogenesis and in response to stresses such as DNA damage or changes in nutrient availability.
Figure 6. Types of alternative polyadenylation.