The John Coffin Lab
Retrovirus - Host Interaction
Our research interests revolve around many different subjects relevant to obtaining a better understanding of the interaction of retroviruses with their host cells and organisms. Over the past few decades, retroviruses have been remarkably rewarding objects for study, and have revealed and illuminated many aspects of biology, including evolution of host-pathogen relationships, mechanisms of disease, including cancer, AIDS, and others, cellular signaling and trafficking, and fundamental mechanisms of evolution.
Avian Retroviruses and the Evolution of Retrovirus - Receptor Interaction
Avian retroviruses are remarkable in that they have been able to evolve fairly recently to use completely different cell surface proteins as receptors, a process which must involve changes in the regions of the envelope protein necessary for binding and entry. Since there is considerable variation in receptor availability among the natural host (the chicken) of these viruses, such adaptability is crucial for their long-term survival. To study the evolution of receptor usage, we have obtained numerous mutations in a small portion of the relevant region, all of which confer on the virus the ability to enter cells independent of receptor binding. We are testing the mechanism of this effect, as well as the hypothesis that such mutations represent the first step in a complex evolutionary pathway toward use of a completely different receptor.
Effects of HIV DNA Integration on Gene Expression
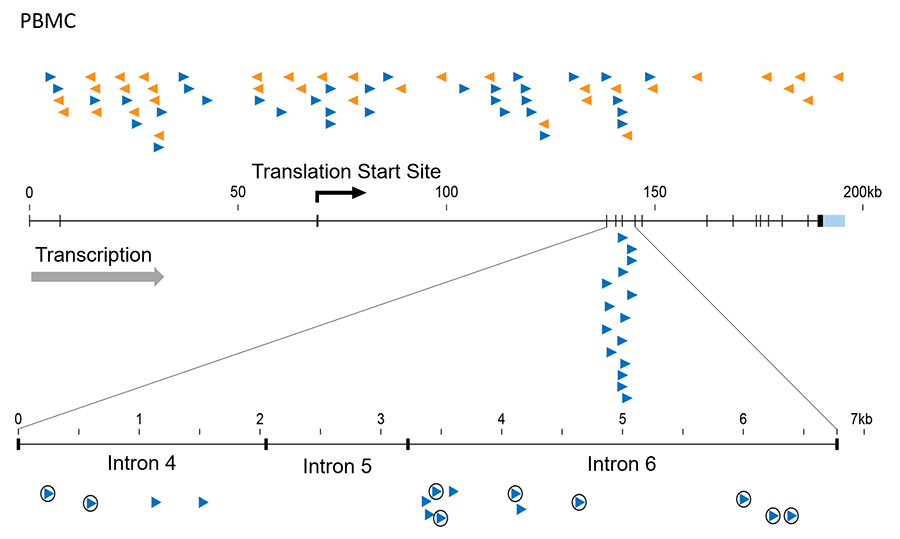
In collaboration with colleagues at NCI-Frederick, we have shown that some HIV infected patients on long-term fully suppressive therapy develop clonal populations of cells with defective HIV proviruses clustered in specific small regions of genes involved in lymphocyte differentiation and cancer, including MKL2 and BACH2. This observation suggests that, like oncogenic retroviruses, HIV DNA integration may influence the expression of genes around the integration site in a way that can modify the host cell growth, survival, and/or differentiation. E affected cells we have seen in patients are quite rare (ca 1 in 10,000), so w have developed an in vitro model based on the use of CRISPR technology to integrate LTRs or defective proviruses at the sites we have observed in patients and then assess the effects on gene expression and cell growth. In related experiments, we are also asking whether natural infection of T cells in culture can lead to selection of cells with proviruses at these or other sites that can also promote cell growth and/or survival.
Figure 1. Integrated HIV proviruses in MKL2 in a patient treated with antiviral therapy for 10 years. The arrowheads and colors indicate the orientation of the provirus. At the top are shown sites of integration in normal PBMCs infected in culture. Note that they are randomly distributed over the whole gene in both orientations. Enlarged at the bottom is a small region (about 12 kb) of MKL2 gene from the treated patient where all 15 proviruses are found and all in the same orientation, and some (circled) are in cells that must have expanded into large clones after infection. This result can only mean that these proviruses affect gene expression to give the cells that carry them a growth or survival advantage.
The "Fossil" Record of Endogenous Proviruses and Virus - Host Evolution
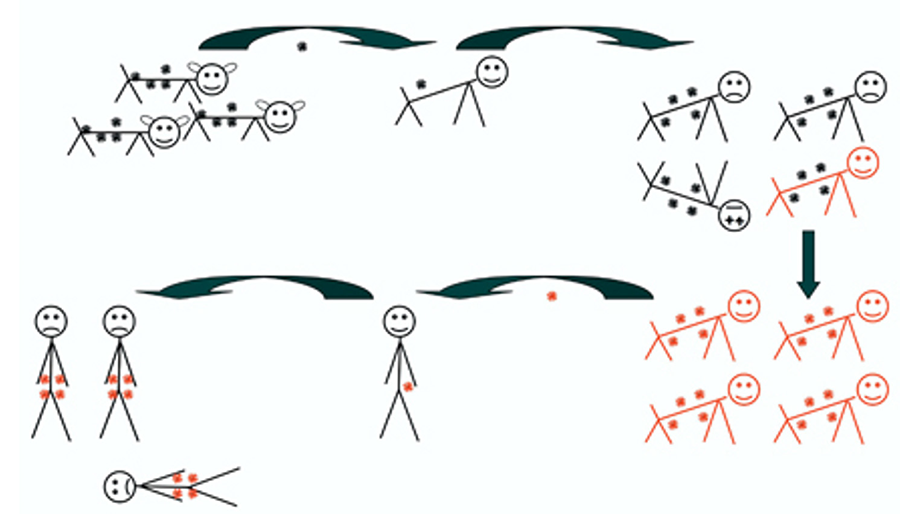
Endogenous proviruses are found in the normal DNA of all vertebrates and many other species. Proviruses and fragments comprise more than 8% of the human genome, several fold more than genes. When a relatively benign virus is transmitted to a new species, it can become much more virulent, leading to adaptation of both host and virus toward a more benign relationship. During this process, some retroviruses can become established in the host genome as endogenous proviruses and transmitted to its descendant species, even long after the infectious virus has become extinct.
Figure 2. The illustration depicts aspects of the co-evolution of endogenous retroviruses with their hosts.
We are pursuing the association of these viruses with human disease, such as the human endogenous virus HERV-K (HML-2) – of which we all carry about 100 proviruses -- with breast cancer and melanoma, by examining the nature, proviral specificity, and consequences of their expression. We are also examining in detail the co evolution of these viruses with both their human and nonhuman primate hosts. We have recently found, for example, that these relics of ancient infection were inserted much more recently into the genomes of gorillas than in humans or chimpanzees, raising the possibility that virus that gave rise to them, although probably extinct in humans, may be still extant as a biologically active, infectious agent in other related species.