The Wai-Leung Ng Lab
The Ng Lab studies the mechanisms used by bacterial pathogens to interact with their own species, with other bacteria, and with the viruses that attack them. We focus on the human pathogen Vibrio cholerae which causes million cases of secretory diarrheal disease cholera worldwide. Our goal is to use our findings to develop new strategies for treating and preventing cholera and other infectious diseases.
The Vibrio cholerae Quorum Sensing System
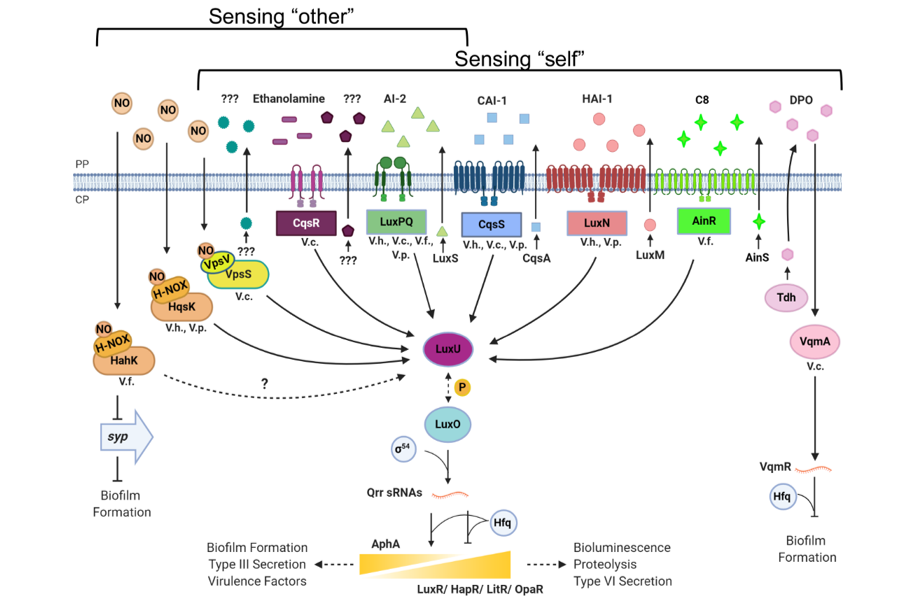
The world is currently in the 7th pandemic of cholera, a devastating diarrheal disease caused by the bacterium Vibrio cholerae. Like many bacterial pathogens, V. cholerae depends on an intricate cell-cell communication system, called Quorum Sensing (QS), to precisely regulate the timing of production of colonization factors and toxins inside the host. We have discovered four signal transduction systems that function in parallel to control QS (Figure 1). These parallel signaling systems sense distinct chemical signals: one signal is exclusively produced by Vibrio species, another signal is produced by many bacteria, and recently we found that a common gut metabolite produced inside the host can be detected by one of these receptors. Surprisingly, any one of these communication pathways appears to be sufficient to foster host colonization. Therefore, the V. cholerae QS system possibly functions to monitor the complexity of the surrounding environments to regulate virulence and biofilm gene expression. We are interested in understanding the molecular mechanisms underpinning signal production and signal detection in these QS pathways. Our goal is to fully understand how and why bacteria perceive multiple parallel sensory inputs to precisely control virulence gene expression in different niches.
Figure 1. Quorum sensing circuits in Vibrio cholerae and other related species.
A novel second messenger signaling pathway important for viral defense
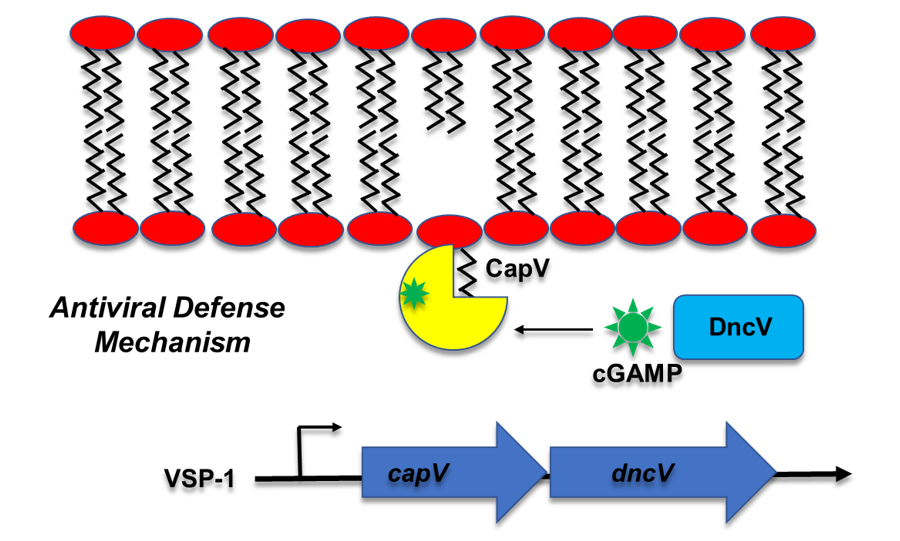
The current 7th pandemic V. cholerae strain is characterized by the possession of many pathogenicity islands (PAIs). These additional genetic elements may improve the fitness of the pathogen. One of the PAIs called VSP-I carry the genes for production of a novel class of second messenger known as cyclic GMP-AMP (cGAMP). Even though cGAMP has been found in other bacteria and is well studied in eukaryotes as a key regulator of innate immune response, how this cyclic dinucleotide regulates its targets in prokaryotes is unclear. We have identified the first protein target of cGAMP, a phospholipase called CapV, in V. cholerae (Figure 2). CapV activated by cGAMP leads to programmed cell lysis which is believed to be an antiviral defense mechanism. We are interested in understanding how cGAMP activates its targets and the role of cGAMP signaling in preventing virus (bacteriophage) attack to the pathogen.
Figure 2. The Vibrio cholerae cGAMP signaling pathway
Interaction between V. cholerae and gut microbiota species
Gut commensal microbes (microbiota) are recognized as an important and potentially tractable host feature that determines host susceptibility to enteric infection. We have identified certain gut microbes that change the virulence and biofilm formation of V. cholerae. We are interested in identifying the mechanisms of cholera-microbiota interactions that alter the virulence of the pathogen to increase our understanding of colonization resistance and host susceptibility.