The Gordon Huggins Lab
Identifying Genes Associated with Human Disease
We believe firmly in the benefit of making primary discoveries of human disease whenever possible in humans. Within the past one to two decades the human genome sequence has been completed and a map of genetic markers suitable for studying disease associations has been established. Both candidate gene and genome-wide association studies are performed in the Huggins laboratory. A particular focus is the identification of genetic contributors to bicuspid aortic valve, hypertrophic Cardiomyopathy, dilated Cardiomyopathy as well as metabolic factors including lipoproteins and inflammatory markers. Recently, we are exploring the use of novel genetic approaches for the comprehensive identification of rare and common coding variants associated with heart muscle disease. These large-scale genetic screens are supported by active collection of DNA and serum samples from patients of the Tufts Medical Center Cardiovascular Center.
Analysis of Hypertrophic Cardiomyopathy
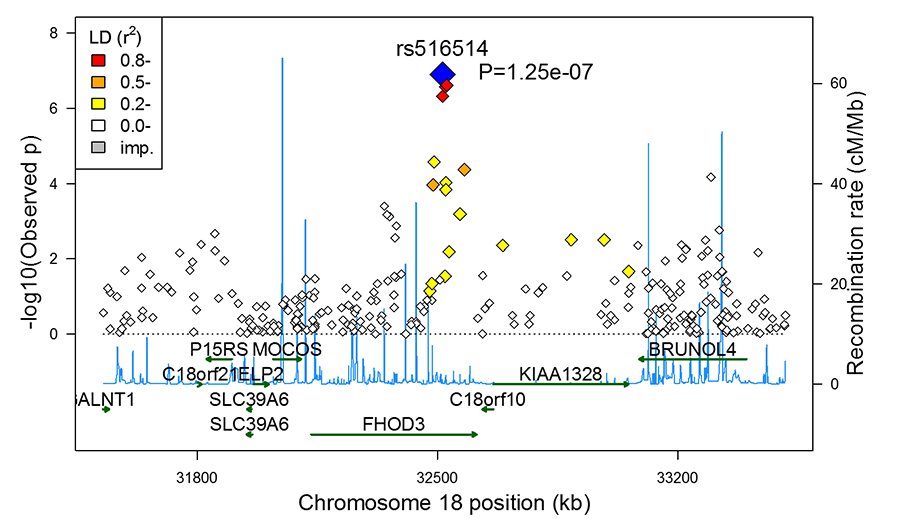
Causative factors of heart muscle disease include environmental insults such as ischemia/infarction, high blood pressure, toxins, drug exposure and atypical genetic changes. Hypertrophic cardiomyopathy (HCM) is an example of a heart muscle disease caused by gene mutations. The Huggins laboratory is analyzing how a member of the form in family of genes contributes to heart muscle disease including HCM (Figure 1).
Figure 1. Genome-wide association analysis identifying a locus in FHOD3 as being associated with hypertrophic cardiomyopathy.
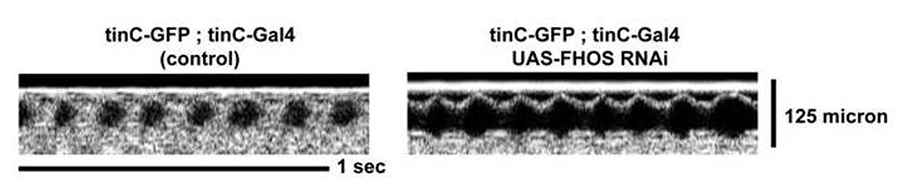
This highly conserved gene is required for normal heart function in flies as measured by optical coherence tomography (Figure 2).
Figure 2. Fly heart contractile function is reduced in flies that have knockdown of fhos, (the fly homologue of FHOD3 (right), compared with control flies.
Calcific Aortic Valve Disease
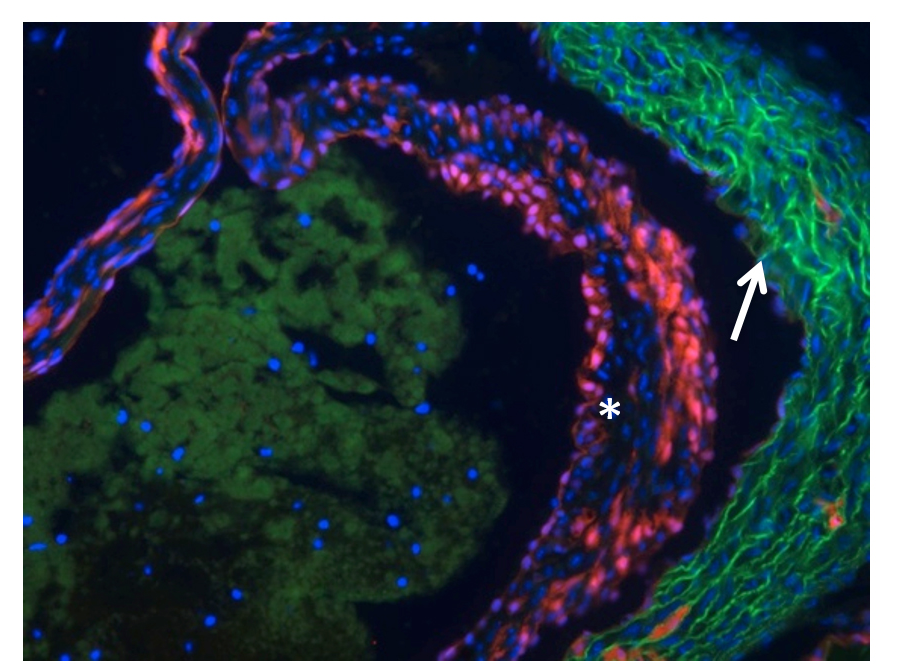
Our laboratory is collaborating with Drs Phil Hinds and Lauren Black on the role of the retinoblastoma protein (pRb) in aortic valve disease. By selectively targeting pRb in the Tie2 lineage (Figure 3) we have developed an age-dependent model of aortic valve disease with which they hope to define the relative contribution of valve cell autonomous effects versus systemic inflammation in the genesis of aortic valve disease.
Figure 3. Demonstration of Tie2Cre-dependent recombination within the aortic valve leaflet of Ai9 mouse indicated by TdTomato fluorescence (*). Autofluorescence is seen in the the aortic wall (indicated by arrow)