The Rui Guo Lab
Combating Epstein Barr Virus (EBV) and Its Health Implications
Epstein Barr Virus (EBV) is the first human tumor virus. It infects over 95% of adults across the globe and is known to cause infectious mononucleosis. Notably, it is linked with around 200,000 cancer cases each year. A stark manifestation of its lethal potential is seen in Africa, where EBV+ Burkitt lymphoma prevails as the leading childhood cancer. Furthermore, EBV is highly associated with various other cancer types like Hodgkin, HIV-associated, and post-transplant lymphomas, as well as nasopharyngeal and gastric carcinomas.
Our lab is dedicated to exploring the complexities of EBV, particularly its pathogenic role in cancer initiation and development. Current treatments for EBV+ cancers do not effectively target the EBV-driven pathways, leading to significant toxicity and limited efficacy. In response to this challenge, our research focuses on targeting vulnerabilities in EBV-driven metabolic and epigenetic processes, aiming to pave the way for new treatments. Ultimately, we strive to integrate CRISPR genome editing, transcriptomics, metabolomics, and proteomics to study interactions between tumor viruses and lymphocytes and epithelial cells.
Approach
To discover new therapeutic targets for EBV associated cancers, we're utilizing a wide range of advanced techniques:
Molecular Biology, Transcriptomics, and Metabolomics
Utilizing RNA sequencing, metabolomics, and isotopic tracing, we aim to delve into the cellular and molecular mechanisms of EBV, exploring how it interacts with and manipulates host's cellular processes for its latency and transformative capability.
Epigenetics;
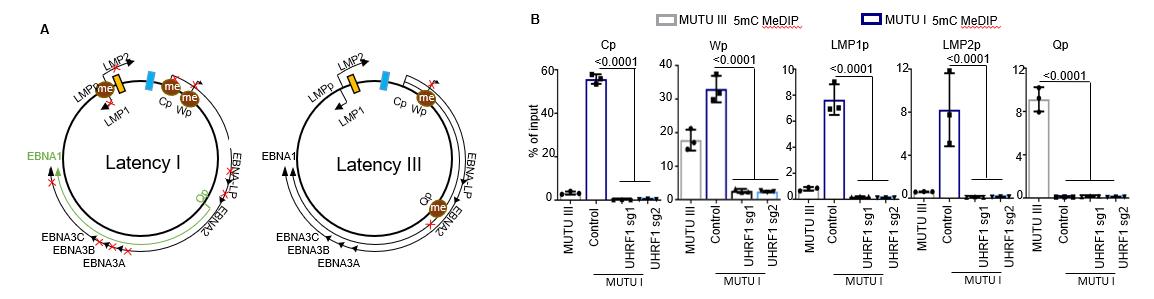
We use ChIP-seq, ATAC-seq, MeDIP-seq and chromatin conformation capture(3C) to investigate how EBV modulates genome hypermethylation and high-dimensional genomic structure, which maintains the virus latency and simultaneously turns off tumor suppressors (Figure 1).
Fig 1. UHRF1 maintains EBV latency promotor methylation (Guo, et al. 2020. Nat. Microbiol. Aug;5(8):1051-1063.doi: 10.1038/s41564-020-0724-y. Abstract ). A, Schematic diagram of EBV latency Cp, Wp and Qp promotor usage in latency I (left) versus latency III (right). Brown circle indicates DNA CpG methylation, red “x” indicates epigenetically blocked transcription. B, MeDIP was performed in MUTU III (gray bars) or in MUTU I (blue bars) expressing control or independent UHRF1 sgRNAs followed by qPCR with primers specific for Cp, LMP1p, LMP2p or Qp. Mean + SEM for n=3 replicates are shown. ****, p < 0.0001.
Genome-wide CRISPR/Cas9 Screening
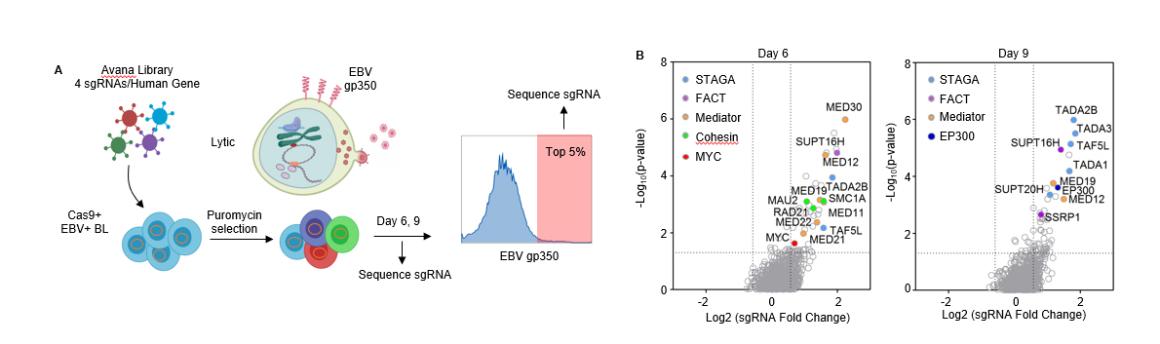
We are using CRISPR technology to identify potential genetic targets in EBV-driven metabolic networks (Figure 2).
Fig. 2. Genome-wide CRISPR Screen for Suppressors of EBV Lytic Reactivation (PMID: 32315601). A, CRISPR/Cas9 screen workflow and screening strategy. Cas9+ P3HR-1 BL-cells were transduced with the Avana sgRNA library at a MOI 0.3, selected with puromycin and sorted for top 5% of gp350+ cells. sgRNA abundances from input versus sorted cells were quantitated. B, Volcano plots showing the -Log10(p-value) statistical significance (y-axis) and Log2 fold-change (x-axis) of sgRNA abundance in input versus Day 6 output (top) or Day 9 output (bottom). Statistical significance was quantitated by STARS analysis of biological screen replicates. Selected screen hit categories are highlighted.
Our Research Revolves around Three Questions
EBV-driven Methyl Group Metabolic Pathways
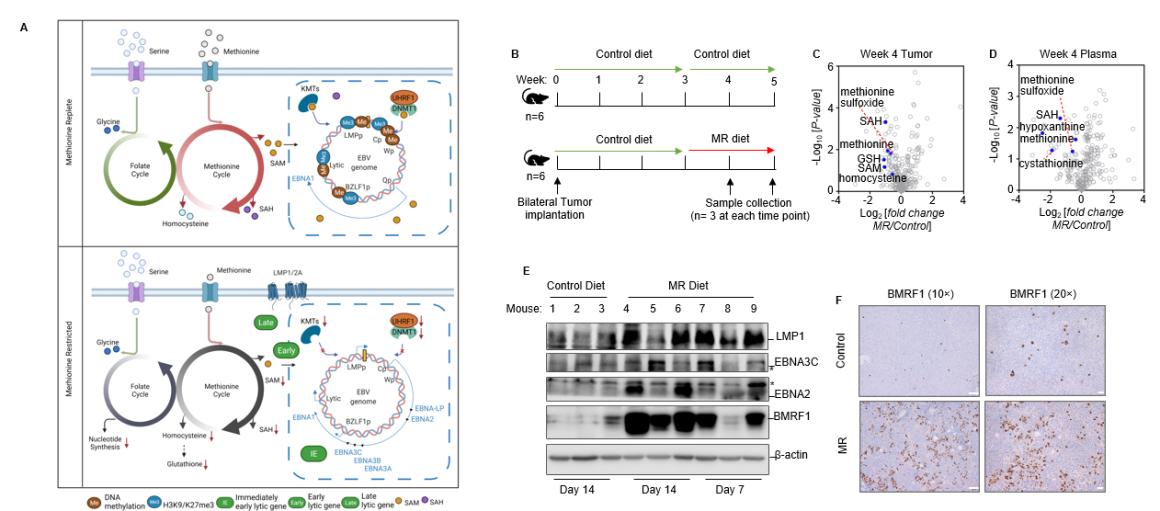
We are investigating how EBV controls methyl group metabolic pathways, leading to genome hypermethylation that keeps the virus latent and suppresses tumor suppressor genes (Figure 3).
Fig. 3. Methionine restriction awakens EBV immunogenicity in Burkitt Lymphoma (PMID: 36070681). A. Schematic model of interconnected folate and methionine metabolism cycle roles in control of the Burkitt B cell EBV epigenome, latent and lytic gene expression. We identified interconnecting methionine and folate cycles as critical for programming the double-stranded DNA EBV epigenome in Burkitt lymphoma cells in support of immunoevasion. Perturbation of methionine or one-carbon metabolism hypomethylates the EBV genome and derepresses immunogenic viral latency and lytic cycle antigens, including in tumor xenografts. B. Schematic of mouse xenograft control versus MR diet intervention. Mice were feed with control diet with 100% methionine or MR diet with 10% methionine. C-D. Volcano plot analysis of -Log10 (p-value) versus Log2 fold-change of metabolite abundances in n=3 tumors (B) or plasma samples (C) collected from mice fed MR vs control diets for 14 days. Methionine cycle metabolites were significantly reduced in tumors and plasma in mice fed with MR diet. E. Immunoblot analysis of whole cell lysates from tumors explanted from control or MR diet fed mice. Blots of EBV proteins and actin are representative of n=3 analyses. F. Immunohistochemical analysis of EBV lytic protein BMRF1 expression in tumors of mice fed control or MR diet for two weeks.
EBV-driven Mitochondrial Specialization
We are exploring how EBV creates specialized mitochondria, which direct energy and biosynthetic pathways, supplying the necessary building blocks to fuel cellular growth and transformation.
EBV-driven Triglyceride Storage and Hydrolysis
We are studying how EBV manages triglyceride storage and hydrolysis, aiding the transformation of B- and epithelial cells.
Summary
Through investigating these questions with our advanced techniques, we are developing a research pathway that seeks to unravel the mysteries of EBV and pave the way for new, effective, and safe treatment options against EBV+ cancers. Our work is driven by the collective expertise of our committed team and an unwavering quest for scientific knowledge and innovation.