The Rob Blanton Lab
Cardiac Remodeling and Heart Failure
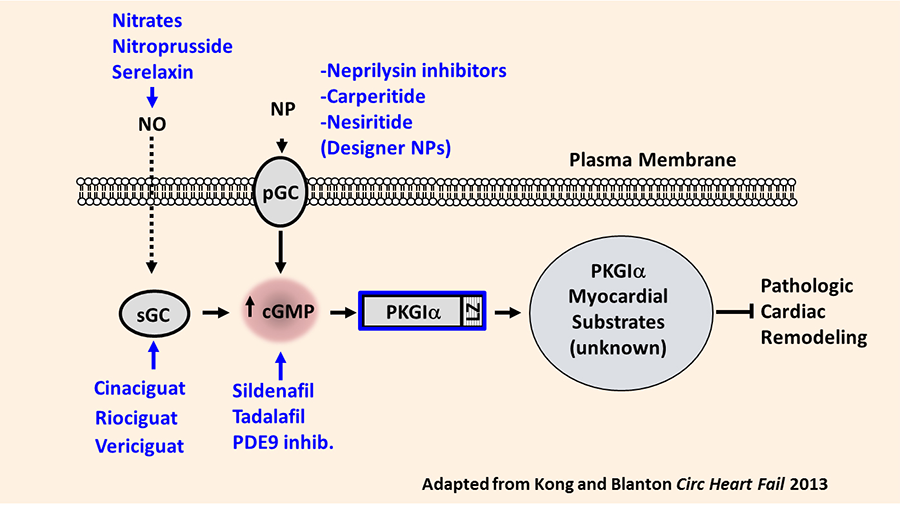
Heart failure remains the leading cause of hospital admission in the US. Our laboratory investigates the basic molecular signaling mechanisms regulating the process of cardiac remodeling: the myocardial structural and functional abnormalities which ultimately cause the heart failure syndrome. We have identified that the cGMP-dependent protein kinase I alpha (PKGIα) opposes pathologic cardiac remodeling through mechanisms which require the PKGIα leucine zipper protein interaction domain. Specific leucine zipper dependent PKGIα substrates mediating this effect remain poorly understood. Understanding these substrates is of clinical and scientific interest since PKGIα activating compounds remain under investigation in patients with heart failure. Our overarching hypothesis holds that myocardial PKGIα kinase substrates function as novel anti-remodeling molecules and therefore represent candidate therapeutic targets in cardiac remodeling and heart failure (Figure 1).
Figure 1. Rationale for exploring PKGIα substrates as novel anti-remodeling molecules. Summary of PKGI activation pathway in cardiovascular tissues. Current available pharmacologic agents targeting specific components of the pathway are shown in blue. Also represented are the hypothesized PKGI substrates in the cardiac myocyte, which may selectively attenuate pathologic cardiac remodeling. NO, nitric oxide; sGC, soluble guanylate cyclase; pGC, NP, natriuretic peptide; pGC, particulate guanylate cyclase (NP receptor); PKGI, protein kinase G I; LZ, leucine zipper domain.
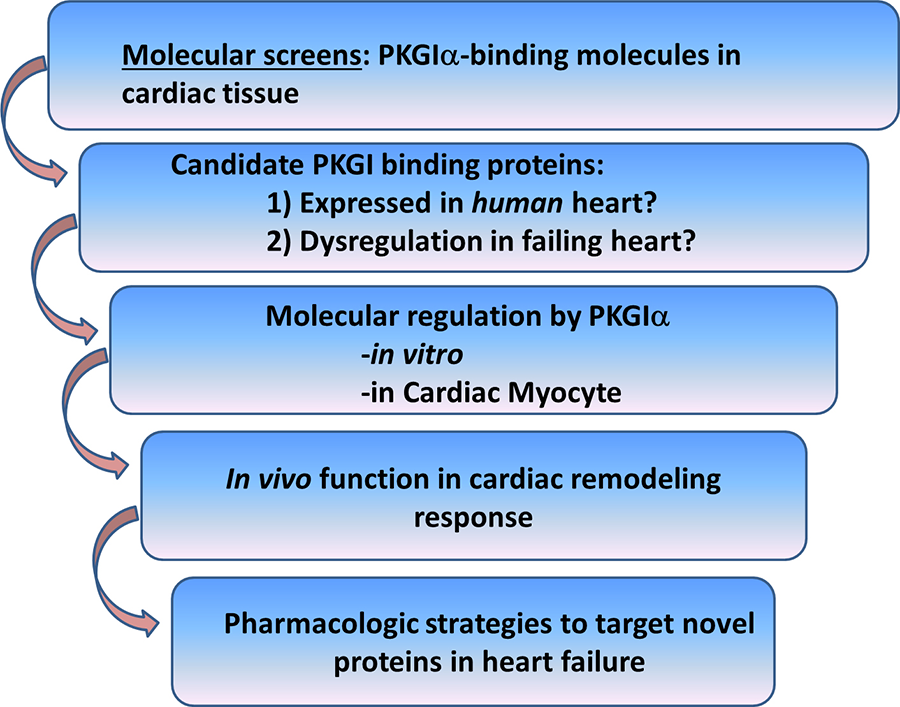
To this end, we have identified a number of novel cardiac PKGIα substrates, and current work in the lab focuses on understanding the role of these molecules in opposing pathological cardiac remodeling. We carry out these studies in vitro, in cultured cardiac myocytes, and in vivo models to determine how these signaling molecules inhibit cardiac remodeling and to what degree these effects require PKGIα (Figure 2).
Figure 2. Discovery strategy for identification of PKGIα cardiac substrates as candidate therapeutic targets for treatment of heart failure.
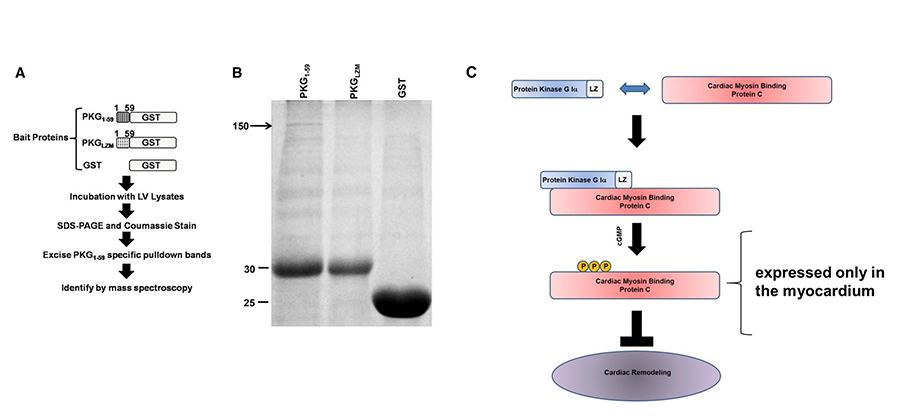
As an example, we recently identified cardiac myosin binding protein-C (cMyBP-C) as a cardiac myocyte-specific PKGIα anti-remodeling substrate (Figure 3). This study supports the notion that PKGIα inhibits cardiac remodeling through protein-protein interactions with LZ-dependent substrates. This has provided rationale for the broader discovery strategy described in Figure 2, in which we are exploring additional novel PKGIα binding molecules as potential therapeutic targets for cardiac remodeling and heart failure. Current projects in the lab range from: 1) performing molecular screens, to 2) studying the regulation and function of PKGIα substrates for their role in the cardiac myocyte and in modulating the cardiac remodeling response in vivo.
Figure 3. Identification of cardiac myosin binding protein-C (cMyBP-C) as a cardiac myocyte-specific PKGIα anti-remodeling substrate through molecular screen for PKGIa-LZ binding proteins. From Thoonen et al, 2015. (A) Outline of screening strategy. GST-fusion proteins were generated containing the PKGIa LZ domain (PKG1-59), the PKGIα mutated LZ domain (PKGLZM), or GST alone. The separate bait proteins were incubated with left ventricular protein lysates, followed by SDS PAGE and Coomassie staining. Protein bands selectively precipitating with PKG1-59 were removed and identified by mass spectroscopy. (B) Representative Coomassie stain from left ventricular protein lysates incubated with GST-fusion proteins. The 150 kDa band visible only in PKG1-59 precipitate (denoted by arrow) was excised and subjected to mass spectroscopy, revealing cMyBP-C as the predominant species. The thick bands between 25 and 30 kDa represent GST fusion proteins. Representative of 3 independent experiments. (C) Model for PKGIα phosphorylation of cMyBP-C on conserved serines leading to attenuation of cardiac remodeling.