The Peter Juo Lab
Synapse Development and Function in C. elegans
Research in the lab is focused on understanding the cell biological mechanisms that regulate glutamatergic synapse development and function. In particular, we are interested in identifying novel genes and mechanisms that regulate the trafficking, localization and abundance of glutamate receptors at synapses. Glutamate is the major excitatory neurotransmitter receptor in the mammalian brain and regulation of glutamate receptor levels at synapses is important during synapse development and for learning and memory. Aberrant glutamatergic synapse development and function are associated with several neurodevelopmental and neuropsychiatric disorders, such as Autism Spectrum Disorders, Alzheimer’s Disease, Schizophrenia, Depression and Epilepsy. Understanding how normal healthy synapses develop and function provides the foundation for discovering what may be altered in various diseases of the nervous system.
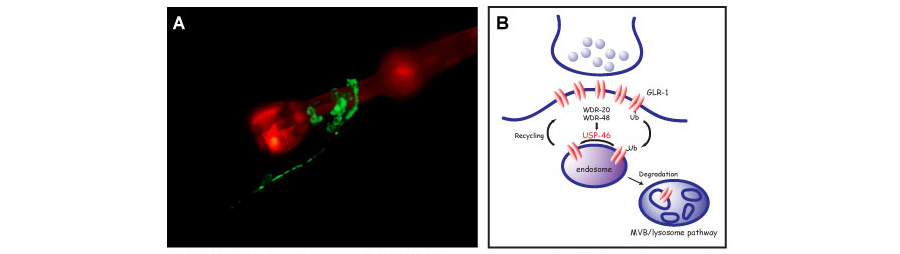
We use a combination of genetic, biochemical, imaging and behavioral approaches to study glutamatergic synapses in vivo in the genetic model organism C. elegans. The nematode C. elegans has several advantages for studying synapse biology, including a compact nervous system with exactly 302 neurons, simple behaviors that can be correlated with a defined wiring diagram, powerful genetic tools for in vivo analysis of gene function, and a transparent cuticle that facilitates fluorescence microscopy (Figure 1A).
Figure 1. (A) Confocal image showing GLR-1::GFP expressing neuronal cell bodies and processes in the nerve ring and anterior ventral nerve cord in the head of the worm. The nerve ring wraps around the pharynx, which is shown expressing the red fluorescent protein mCherry. The worm is oriented ventral side down with its anterior to the top right of the image. (B) Model of USP-46 at the synapse. WDR-20 and WDR-48 activate the deubiquitinating enzyme USP-46 to remove ubiquitin from GLR-1 and protect the receptor from degradation in the lysosome.
We have used genetic screens to identify several novel genes that regulate the trafficking and turnover of the glutamate receptor GLR-1 at synapses in C. elegans. For example, one project in the lab is focused on understanding how the ubiquitin system regulates synaptic GLR-1 levels. Like phosphorylation, ubiquitin-modification of proteins has emerged as a widely used posttranslational modification to regulate diverse cell biological functions. Studies in C. elegans were the first to show that glutamate receptors are regulated by ubiquitin, which promotes GLR-1 internalization and degradation in the lysosome. We used an RNAi screen to identify the conserved deubiquitinating enzyme (DUB) USP-46, as the first DUB to regulate glutamate receptor trafficking and stability. Loss-of-function mutations in usp-46 result in decreased levels of GLR-1 at synapses and corresponding defects in glutamatergic behavior. We are currently studying how two WD40-domain proteins, WDR-20 and WDR-48, interact with and regulate USP-46 to control GLR-1 levels at the synpase (Figure 1B).