The Malavika Raman Lab
Recognition of Ubiquitin-Modified Substrates by VCP adaptor Complexes
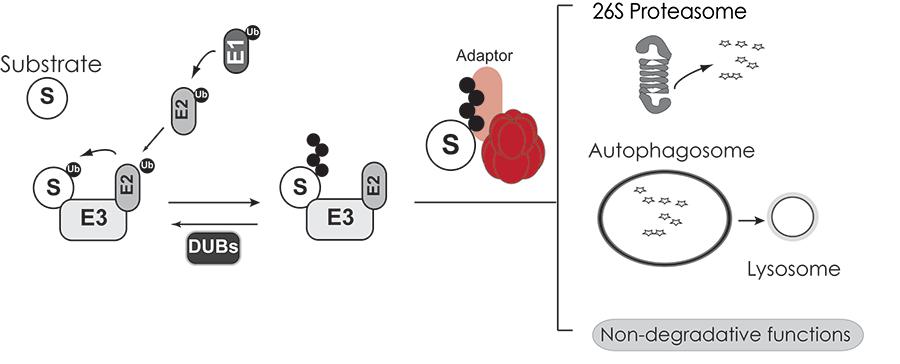
VCP has emerged as a central regulator of protein quality control and ubiquitin-mediated signaling events. This abundant cellular chaperone performs critical functions in diverse cellular programs such as ER associated degradation, autophagy, DNA damage responses and chromatin re-modeling. ATP hydrolysis by VCP generates mechanical forces needed for remodeling client protein complexes and unfolding ubiquitinated substrates. The ability to impact these diverse processes is brought about by targeting VCP to specific cellular structures and substrates via VCP-specific adaptor proteins (Figure 1).
Figure 1. Substrates modified with ubiquitin chains are recognized by VCP AAA-ATPase and associated adaptor proteins. Substrates are subsequently delivered to a variety of protein triage pathways.
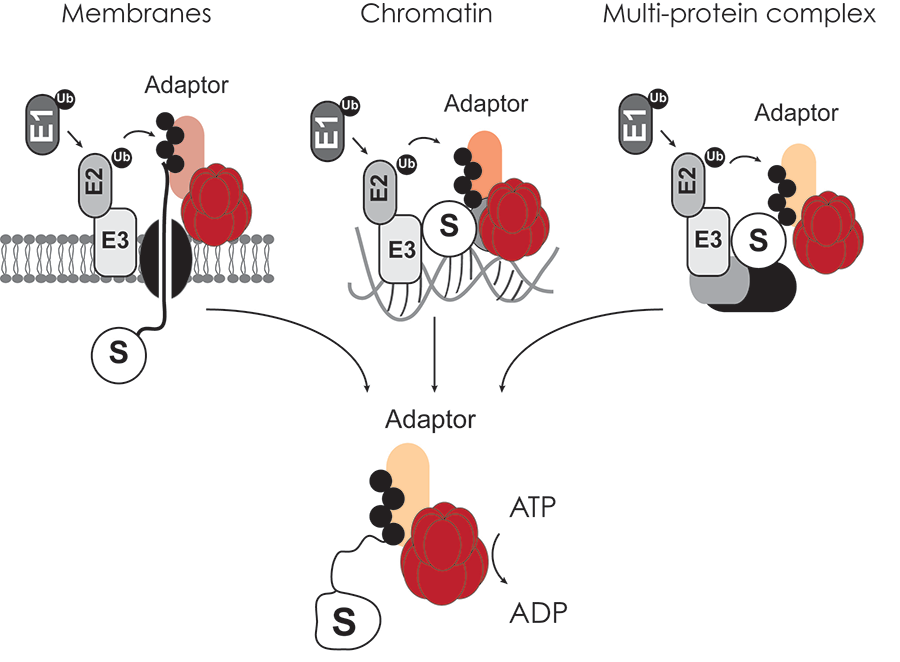
Our recent work interrogating the VCP-adaptor interaction landscape using proteomics suggests that VCP-specific adaptors target VCP to diverse cellular organelles and substrates therein. We aim to pursue these complexes in greater detail to gain a more mechanistic understanding of VCP function within cells. In particular we are interested in determining how distinct VCP-UBXD adaptors interface with E3 ligases, substrates and specific cellular organelles (Figure 2).
Figure 2. The VCP AAA-ATPase functions on diverse cellular targets modified with ubiquitin. ATP hydrolysis by VCP aids in remodeling complexes and delivering ubiquitinated substrates to the proteasome for degradation
Cellular triage pathways impacted by VCP
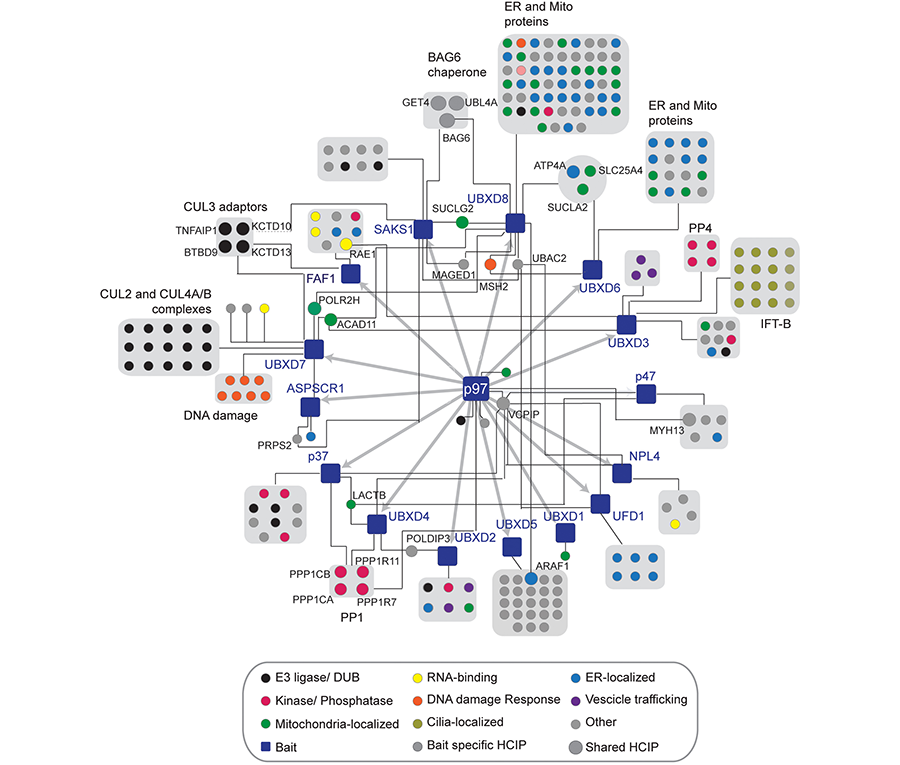
The identification of VCP mutations in individuals with neurodegenerative disorders such as Inclusion Body Myopathy, Paget’s disease of the bone and Frontotemporal Dementia (IBMPFD) and Amyotrophic Lateral Sclerosis (ALS) highlights the importance of tightly regulating VCP function within cells. These disorders are characterized by inclusion bodies and rimmed vacuoles containing ubiquitinated aggregates suggestive of deficits in both the UPS and autophagy. Strikingly, the majority of mutations map to the region in VCP where a class of adaptors that harbor a ubiquitin X domain (UBXD) bind. It is widely thought that mutations may impact the ability of VCP to bind to distinct classes of adaptors thereby skewing its activity towards certain pathways. However, we do not yet have a full understanding of how the numerous mutations globally impact VCP-dependent processes. A major goal of our lab is to understand how distinct VCP-UBXD modules function in triage pathways implicated in neurodegenerative disorders using a combination of genetic, proteomic and biochemical strategies (Figure 3).
Figure 3. The cellular interaction landscape of VCP-UBXD adaptor complexes.
Global Proteomic Profiling of VCP cellular targets
While we appreciate the importance of VCP in many cellular pathways, we have little knowledge of the substrates it targets with in those pathways. Small molecule inhibitors of VCP are currently in clinical trials for certain cancers yet the specific substrates that are stabilized thereby leading to cytotoxicity are not known. We aim to identify the cellular cohort of proteins that rely on VCP function using quantitative proteomic strategies. In addition, using CRISPR-based gene editing technologies we would like determine cohorts of VCP substrates that are dependent on specific classes of adaptors. Ultimately these studies will determine if and how these VCP-adaptor-substrate substrates are mis-regulated in human disorders (Figure 3).