The Linc Sonenshein Lab
A Global Regulator of Metabolism, Nutritional Adaptation, Sporulation and Virulence in Gram-Positive Bacteria
The principal focus of our lab is to understand how bacteria adapt to changes in nutritional availability. The exhaustion of favored nutrients is a universal signal for bacteria to swim to new locations, to synthesize enzymes that degrade macromolecules and transporters that bring the degradation products into the cell, to turn on intracellular degradative pathways and to secrete antibiotics that kill competitor bacteria in the local environment. All of these responses are designed to help bacteria restore their growth potential by using alternative nutritional sources. Some bacteria, especially the Gram-positive Bacillus and Clostridium species, have a fall-back option if these attempts to restore growth fail; that is, they can induce spore formation, thereby converting the growing cell into a highly stable, dormant form that can survive for a very long time even in very harsh environments. Some species of Gram-positive bacteria, whether spore-forming or not, are pathogenic for humans or other animals. In most cases, these bacteria do not express their virulence proteins, especially those that act as toxins, all the time but only when the bacteria are nutritionally stressed. Through our studies of nutritional adaptation in Gram-positive bacteria, we discovered a global regulatory protein called CodY that is ubiquitous in these bacteria and controls key aspects of metabolism, adaptation to nutritional stress, sporulation and virulence.
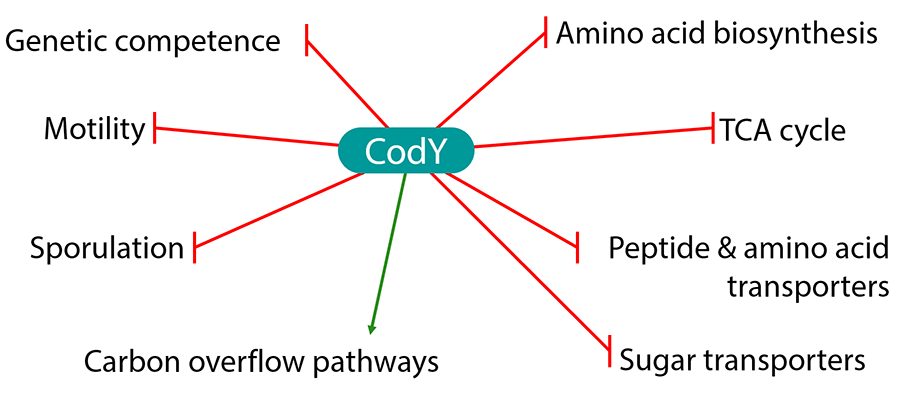
CodY is a dimeric protein whose ability to bind to specific sites on DNA is activated by its interaction with two types of metabolites - - the nucleotide GTP and the amino acids isoleucine or leucine or valine. We have defined the amino acid binding site within CodY both by genetic experiments and, in collaboration with the group of Dr. A. J. Wilkinson (University of York, UK), by structural analysis. In the non-pathogenic bacterium Bacillus subtilis, CodY activates transcription of key metabolic genes needed for active growth and represses the expression of genes needed for adaptation to nutritional limitation.
Figure 1. Gene regulation by CodY in Bacillus subtilis.
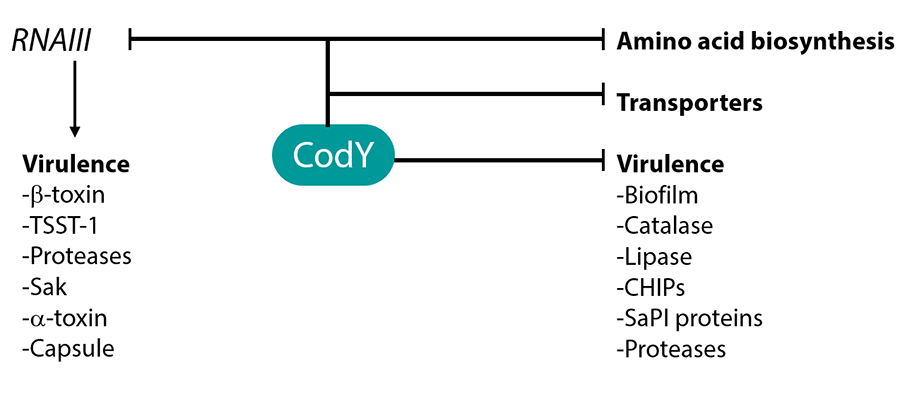
In Staphylococcus aureus, the causative agent of many diseases (boils, toxic shock syndrome, pneumonia, and osteomyelitis), CodY has additional roles beyond regulating metabolism and nutritional adaptation. That is, CodY represses the synthesis of the many toxins that are the primary causative agents of disease. It does so by binding directly to promoter sites of some of the toxin genes, but also controls virulence indirectly by repressing the synthesis of RNAIII, a regulatory RNA that is the direct controller of many virulence genes.
Figure 2. Direct and indirect roles of CodY in the regulation of Staphylococcus aureus virulence genes.
In Listeria monocytogenes, by contrast, CodY may serve as a positive regulator of virulence. Whether it does so directly or indirectly is an unresolved question and a current topic of detailed analysis in our laboratory.
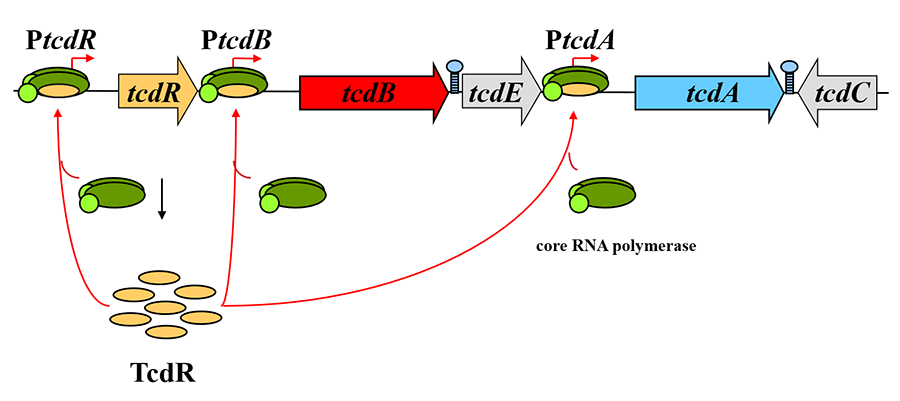
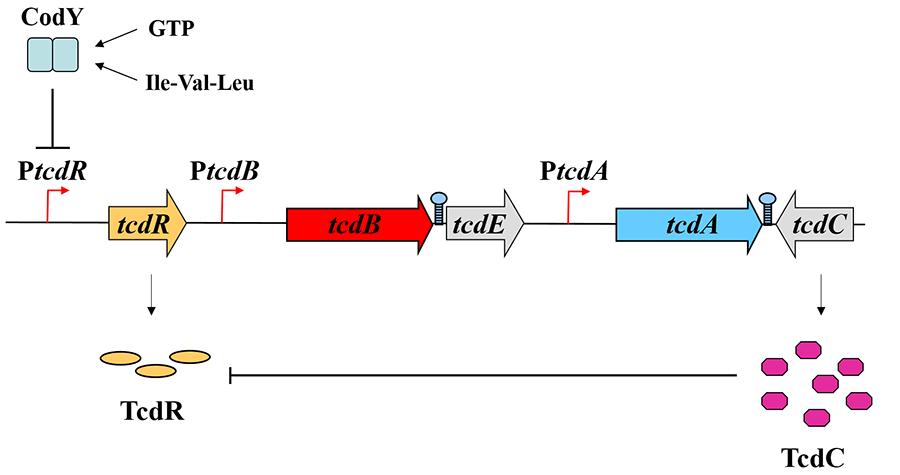
Clostridium difficile is the most frequent cause of antibiotic-associated gastroenteritis among hospitalized patients. This potentially devastating disease has been very difficult to treat, so new approaches to prevention and therapy need to be identified. In this bacterium, synthesis of the two major toxin proteins (TcdA and TcdB) requires the activity of TcdR, an alternative sigma factor (promoter-recognizing subunit) of RNA polymerase. The synthesis of TcdR is restricted to nutritionally limited cells because it is strongly repressed by CodY in rapidly growing cells.
Figure 3. TcdR is a sigma factor for toxin gene transcription.
A second regulatory protein, TcdC, was shown by our collaborator, Dr. Bruno Dupuy (Institut Pasteur, France), to inhibit the activity of TcdR. This dual regulation of TcdR synthesis and activity prevents toxin gene expression during rapid growth.
Figure 4. A model for toxin regulation in C. difficile under conditions of nutrient excess
A key question that arises is whether CodY could be a useful target for novel anti-infective agents. For instance, we could imagine compounds that are sufficiently similar to isoleucine that they activate CodY, but sufficiently different that they would not be metabolized by the bacteria. If patients were fed such compounds, the C. difficile cells in their intestinal tracts might maintain CodY in its active (tcdR-repressing) state even when the cells are nutritionally limited, thereby reducing the virulence of the bacteria. The same compounds might also reduce the virulence of S. aureus and all other Gram-positive bacteria in which CodY represses important virulence genes.
We are also studying the conditions that lead to germination of C. difficile spores in the intestinal tract, since spore germination is the first step in infection. We have found that C. difficile spores have the unique property of sensing bile acids as germinants. Moreover, whereas some bile acids stimulate germination, others are competitive inhibitors of germination. We have been designing and testing non-metabolizable analogs of the inhibitory bile acids as potential anti-infectives.