The James Baleja Lab
The goal of my research is to understand the mechanisms by how cells alter their metabolism in response to an external perturbation, such as the loss of a gene or lack of blood supply to a tissue. Our approach uses biophysical and biochemical methods such as NMR spectroscopy.
Characterization of small molecules in controlling cell fate
Our lab uses and develops metabolomics technologies to examine biological problems in a translational medicine context. Metabolomics is a field of systems biology centered on the study of small biological molecules in biological fluids and tissues. Recent research suggests that analysis of metabolite concentrations in living systems is useful in disease diagnosis, prognosis, and predicting drug efficacy in a personalized medicine context.
The metabolome is the set of all metabolites in a tissue, or cell line, growth media, or other biofluid. NMR does not require separation prior to measurement, the sample can be recovered for further analyses, and the method is highly reproducible. NMR is also the basis for magnetic resonance spectroscopy, a procedure that can be added to MRI procedures that provides a non-invasive (without biopsy) biochemical readout of a tissue or organ.
The levels of many metabolites change in response to the changing needs of a cell. For example, many tumors show changes called the Warburg effect, and are characterized by higher glycolytic flux with increased biosynthesis of amino acids, lipids, and nucleic acids to provide for the increased proliferation demands of a tumor. In addition changes occur in the way that cells communicate with each other by the secretion of different metabolites. These changes in metabolites will also change in response to chemotherapy, often before structural changes in tissue are observed. Thus, metabolite levels may provide a valuable biomarker for treatment efficacy. Note altered metabolic pathways include high production of 2-hydroxyglutarate in many glioblastomas and the metabolic dependency on asparagine in acute lymphoblastic leukemia (ALL). Unique metabolic addictions may exist for many types of cancer, and more broadly, many different types of disease.
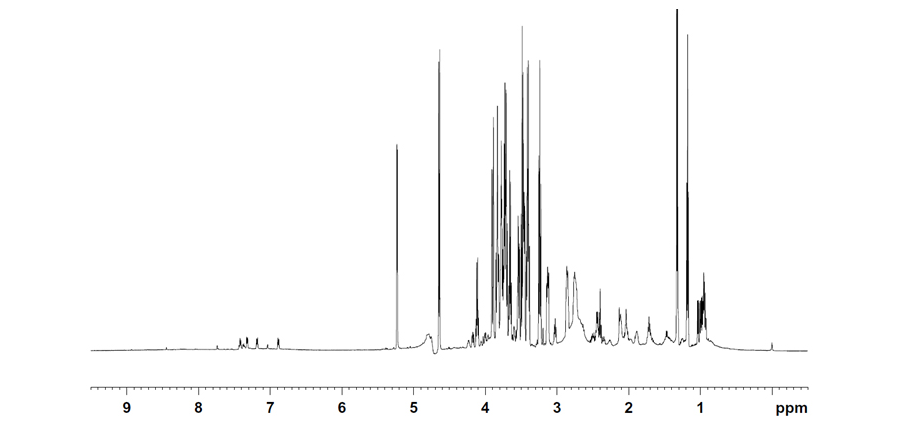
Figure 1. An example of a 1H NMR spectrum illustrating the complexity of molecules present and secreted into growth medium. The ppm scale indicates the resonance frequency that identifies a molecule; concentration is proportional to the height of the peak.
Role of the EH Domain in Endocytosis and Cellular Proliferation
We also are interested in how proteins transmit signals from biological membranes, such as the EH domain-containing proteins. The movement of molecules to and from the surface of a cell is fundamental to many biological processes including glucose uptake, fertilization, cholesterol metabolism, and cellular signaling. These vesicle-mediated sorting mechanisms are mediated in part by interactions between proteins containing EH domains and proteins containing asparagine-proline-phenylalanine (NPF) sequences. The human genome encodes 17 EH domains contained in 11 proteins; about twenty proteins contain NPF motifs that are available for interaction. The gap in knowledge regarding the most likely EH domain/NPF partners is a problem as it prevents us from understanding the molecular interactions that guide the movement of proteins within a cell. For example, the EHD1 protein functions in the recycling of vesicles and thus plays an important role in cellular physiology by directing proteins such as the 1-integrins back to the cell surface as well as controlling the level of the GLUT4-glucose transporter present on the cell surface. Therefore, EH domain containing proteins affect communication of cells with the extracellular matrix, and the homeostasis of glucose and cholesterol within the cell.
Various complexes with EH domains are being characterized to understand specificity at the domain/NPF interface (Figure 2). Our data suggest new protein-protein partners that are being verified using co-localization, GST-pulldown, and coimmunoprecipitation experiments. In addition, the data have provided a basis for the design of specific inhibitors that are being developed and characterized.)
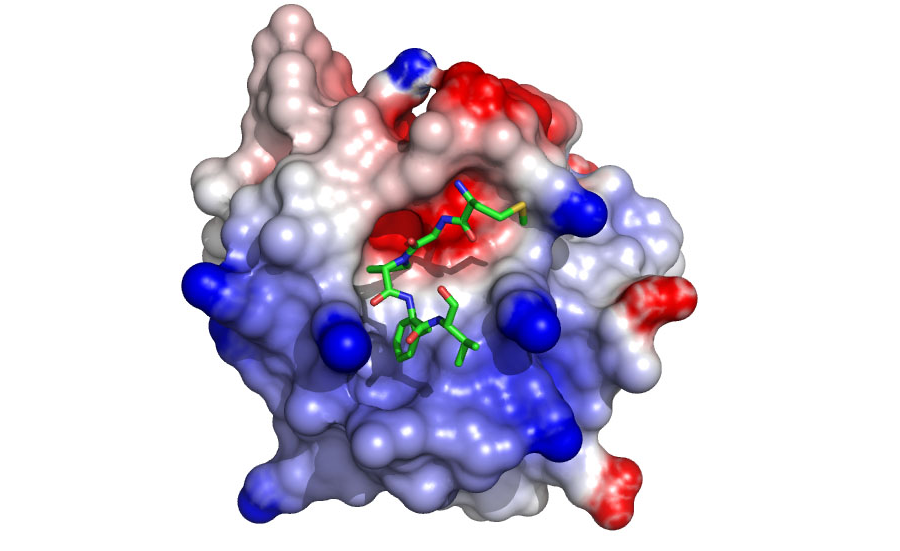
Figure 2. A model of the EHD1 EH domain bound to an inhibitory peptide. The model predicts that the domain uses positively charged residues (in blue) to contact negatively charged residues at the C-terminus of the NPF motif (Henry et al 2010)