The Andrew Greenberg Lab
Our laboratory, called the Obesity Metabolism Laboratory has a long-standing interest in adipocyte and lipid metabolism, nutrition, inflammation, obesity and its metabolic and inflammatory complications. During his postdoctoral fellowship, Dr. Greenberg was the co-discoverer of the first lipid droplet associated protein, adipocyte perilipin (plin1) and initially focused his laboratory’s research on elucidating the role of lipid droplet proteins in regulating cellular and systemic metabolism.
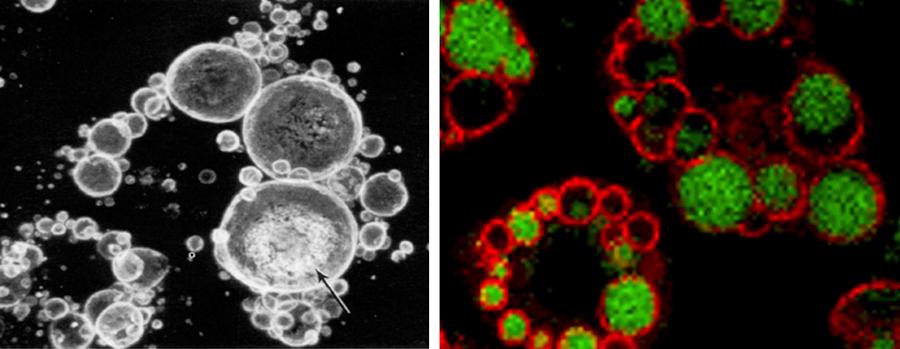
Figure 1. A confocal image of immunostained perilipin around lipid droplets is shown in the left panel. The arrow indicates a lipid droplet. An immunofluorescent staining of perilipin (red) binding to lipid droplets (green) is shown in the right panel.
The laboratory was also the first to demonstrate that adipocytes synthesize and secrete the inflammatory cytokine, interleukin 6. Of additional significance, the laboratory discovered increased rates of adipocyte death in obese mice and humans and this paper is now cited by over 2000 other papers. In obesity, Immune cells surrounding the remnant lipid droplets from dead adipocytes serve as an inflammatory nidus releasing cytokines and metabolites that alters adipocyte metabolism and promotes insulin resistance and hepatic steatosis.
Our laboratory has generated and continues to generate a number of genetically modified mice to determine the tissue specific role of genes in the development of diet-induced obesity and associated metabolic complications such as insulin resistance and hepatic steatosis. We are currently investigating the roles of specific genes in white, brown, and beige adipocytes, macrophages, and intestine in the development of diet-induced obesity and its metabolic complications.
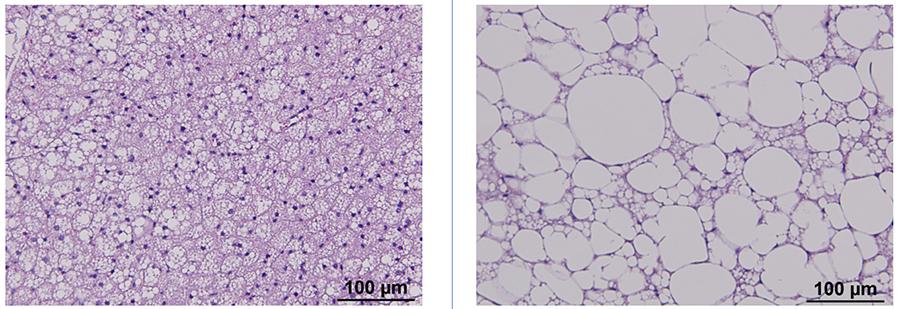
Figure 2. Adipose tissue from lean (left) and obese (right) mice stained with hematoxylin and eosin.
We are also currently using our mouse models to investigate aging-associated alterations in metabolism and atherosclerosis. Our research utilizes a number of techniques including RNA seq, single cell RNA seq, FACS analysis, histology, metabolic chambers and MRI for analysis of mouse energy expenditure and body composition, microbiome analysis, gene and protein expression as well as various microscopic techniques.