The Alexander Poltorak Lab
Genetic Analysis of DNA Responses
Recognition of cytosolic DNA initiates host responses to invading DNA-pathogens as well as to self-DNA that leaks into the cytosol as a result of mitochondrial stresses. In both cases, the responses are mediated by STING (Stimulator of Interferon Genes, encoded by TMEM173) and lead to production of type I IFN, a critical mediator of host responses in pathology and homeostasis. Mutations in humans in DNA-processing enzymes such as TREX1, IFIH1, RNASEH2B, USP18 lead to increased cytosolic DNA, excessive IFN-signaling, and severe, often lethal, pathologies, emphasizing the detrimental impact of excessive amounts of IFN.
In some cases mutations in IFIH1 and TREX1 do not result in a high IFN-signature but still lead to immunopathologies usually associated with excessive IFN production. This suggests additional IFN-independent, potentially pathogenic responses to cytosolic DNA. Further evidence for the complexity of DNA-responses comes from the high prevalence (17% worldwide) of the loss-of-function (LOF) dominantly inherited allele of the TMEM173. This implies that there must be other pathways that can complement or act in lieu of IFN. Thus, defects in DNA-sensing can lead to pathology by mechanisms other than dysregulation of IFN. Taken together, these data suggest that some alternative DNA sensing pathways must have evolved to complement IFN production.
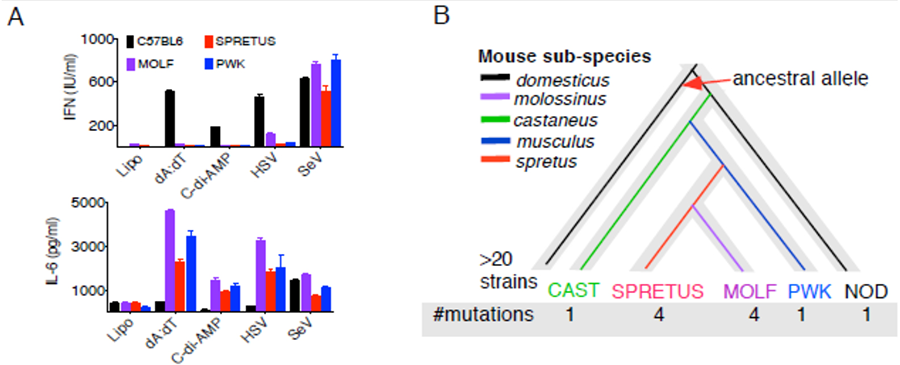
In support of this model, we recently characterized DNA-responses in wild-derived mice MOLF and observed both in vivo and in vitro that MOLF mice respond to cytosolic DNA or DNA viruses by producing massive amounts of IL-6 but low amounts of IFN when compared to C57BL6 (B6), Fig. 1A. Low IFN in MOLF mice is conferred by a hypomorphic allele of Sting, whereas high IL-6 - by several loci including Sting. Similar to MOLF, representatives of M.m.musculus, M.m.molossinus, and M.spretus wild derived mouse sub-species also exhibited low IFN and high IL-6 in response to DNA viruses (see Fig. 1A). Because STING is the main mediator of DNA-responses, we compared STING haplotype and found it highly divergent in all of the wild-derived strains (Fig. 1B). In contrast, more than 20 classical inbred strains of M.m.domesticus lineage have identical STING haplotype (Fig. 1B, black line) thus confirming genetic redundancy of the classical inbred mouse strains. Collectively, despite being inbred, wild derived mice represent significantly more diverse population of wild progenitors than classical inbred strains thereby challenging the current model that has production of IFN as the main outcome of DNA-responses.
Figure 1. STING-mediated DNA-responses in wild-derived mice are biased towards IL-6 (A) – Peritoneal macrophages from M.m.musculus, M.m.molossinus, M.spretus strains of mice produce IL-6 instead of IFN in response to dsDNA (dA:dT), cyclic di-nucleotides (c-di-AMP), DNA-viruses (HSV – Herpes Simplex Virus) but not RNA-viruses (SeV – Sendai virus). Production of cytokines in cell supernatants was measured after 16 hours of treatment with DNA-homologues or viruses. (B) - Molecular evolution of STING protein during sub-speciation of mice. Phylogenetic tree showing relationships between 5 different mouse subspecies; individual strains for each subspecies are presented. New lineages arising from new polymorphisms in the gene are drawn in different colors. The branching of alleles is based on the order of arising of novel mutations. # mutations - number of mutations in each strain, with only unique mutations accounted for. SPRETUS and MOLF have 3 unique and 1 common mutations.
Accordingly, we are trying to identify MOLF-specific components of STING-mediated signaling that confer high levels of IL-6 despite low inducible IFN. This line of inquiry involves:
- Investigating the role of DNA sensors such as cGAS, IFI16, DDX41, AIM2, and STING in DNA-responses in MOLF;
- Investigating the spatial requirements of STING for activation of NF-kB and IFN-signaling;
- Identification of the genes regulating responses to cytosolic DNA in MOLF via forward genetic analysis.
Regulation of Cell Death by Tonic IFN Signaling
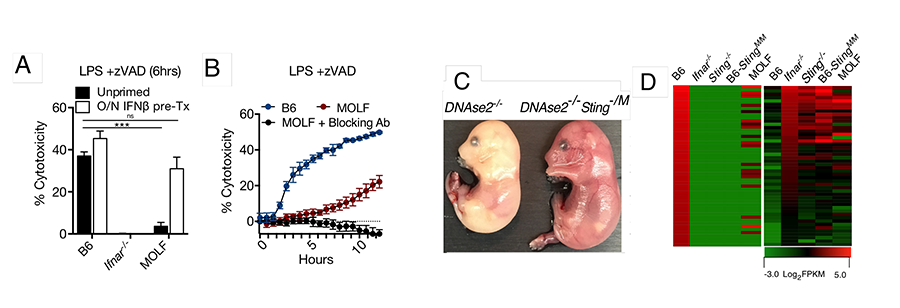
Further investigation of IFN-signaling in MOLF mice has challenged another dogma. That is, we observed that constitutive (tonic) rather than inducible IFN is crucial in initiation of necroptosis. Although basal IFN signaling has been observed since the 1980s, the scope of its function has been purely speculative with no causative link provided. We also established that MOLF macrophages, by the virtue of their hypomorphic Sting allele and therefore weak tonic IFN production are resistant to necroptosis (Fig 2A). Monitoring the cell death in time revealed that MOLF BMDMs exhibit slow necroptosis kinetics and IFNAR blockade during LZ stimulation prevented this late time point sensitization to necroptosis in MOLF macrophages (Fig 2B). Finally, using the MOLF allele of Sting, we rescued the DNase2-KO embryonic lethality (Fig. 2C), which otherwise is rescued by deletion of Sting and also suggested that other than Sting genes confer lethal responses to DNA in MOLF.
Figure 2. MOLF is defective in tonic IFN-signaling and resistant to necroptosis. (A) PropidiumIodide (PI) incorporation 6 hrs following LPS/zVAD (LZ) treatment with or without IFN-pretreatment; (B) PI incorporation during 12 hrs of stimulation with LZ with and without 1 hr treatment with IFNAR blocking Abs; (C) Macropscopic view of 15.5 days old embryos with the indicated haplotype on top; (D) RNA sequencing of rested BMDMs from indicated strains showing genes lower (left) or higher (right) compared to B6 BMDMs.
To look for possible IFN-induced candidates in the necroptosis pathway, we compared the gene expression profile of resting B6 bone marrow-derived macrophages (BMDMs) with BMDMs from Ifnar-/-, Sting-/-, MOLF, and B6-StingM/M using RNA sequencing. We identified approximately 80 genes that are significantly down regulated in resting Ifnar-/-, Sting-/-, MOLF, and B6-StingM/M BMDMs when compared to B6 BMDMs (Fig 2D). One or more of these genes may play a role as an activator of necroptosis that requires constitutive IFN for expression. Additionally, we found approximately 50 genes that are significantly up-regulated in the absence of constitutive IFN (Fig 2D), which would be consistent with one or more of these genes encoding an inhibitor of necroptosis. These data provide support for identification of these effectors of necroptosis using genome-wide screen and a library of lentiviruses harboring more that 125 thousand guided RNAs.