The Aaron Mendez Lab
Identifying and Exploiting Structural Weak Points in Viral Replication Factories
Our lab studies the mechanism by which viruses maximize their replicative output. A strategy that many viruses use to maximize replicative output is the formation of replication compartments. The establishment of viral replication compartments is the essential process during viral replication. These compartments contain all the replicative output of the virus and act like scaffold for the organization of critical replication machinery. Furthermore, replication compartments shield the virus from the human innate immune response and concentrate host factors needed for replication.
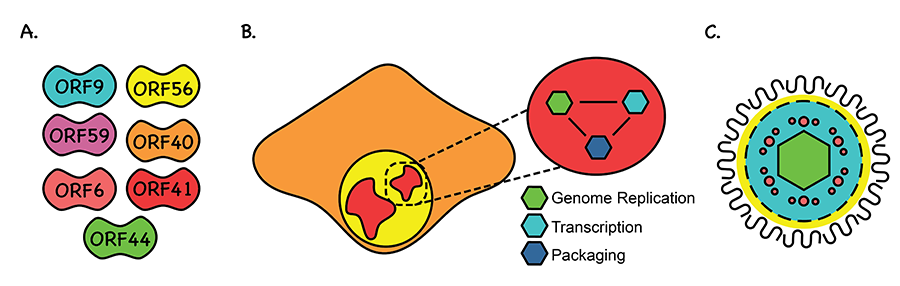
Figure 1. A. There are seven proteins that are essential for the formation of replication compartments, all of which have critical rolls in the replication of KSHV. B. Replication compartments form amorphous structures reminiscent of biomolecular condensates (depicted in red) during early phases of KSHV lytic reactivation. Replication compartments house all the replicative steps of the virus. These include genome replication (green), viral transcription (teal) and packaging (blue). Viral replication compartments are thought to be needed to organize all three of these essential processes. C. The proper assembly of viral replication compartments and organization of the viral replication apparatus is need for efficient propagation of KSHV.
Unfortunately, these sites are difficult to study do to their essential nature during replication. Viral replication compartments are also highly dynamic expanding during the course of replication. A reason for this dynamic nature is that these sites form biomolecular condensate like properties. Historically, biomolecular condensates have been difficult to study due to their multivalent interactions with proteins and nucleic acids. As a result, we have a limited understanding of the key molecular drivers of replication compartment formation and dynamics. Furthermore, how viral replication compartments influence organization of viral machinery, and defend again host innate immune responses is still unresolved.
Our group is nicely positioned to tackle these questions. We are taking a three-prong approach leveraging our expertise in molecular virology, biochemistry, and chemical genetics. First, we have built methods to track replication compartment formation and dynamics using Kaposi’s sarcoma-associated herpesviruses as a model system. Second, we have also generated reconstituted systems of core KSHV replication compartment components. Lastly, we are using chemical genetic approaches to dissect replication compartment assembly.
Our work will provide the foundation for the identification of novel linchpin interactions of viral replication compartments. These site can then be exploited using chemical probes to alter viral replication compartment formation. Furthermore, the tools and methods generated by our work will provide our field with new approaches to study viral replication assemblies.